(Vreeswijk & Sompolinsky, 1996) E/I balanced network
Overviews
Van Vreeswijk and Sompolinsky proposed E-I balanced network in 1996 to explain the temporally irregular spiking patterns. They suggested that the temporal variability may originated from the balance between excitatory and inhibitory inputs.
There are \(N_E\) excitatory neurons and \(N_I\) inbibitory neurons.
An important feature of the network is random and sparse connectivity. Connections between neurons \(K\) meets \(1 << K << N_E\).
Implementations
[1]:
import brainpy as bp
Dynamic of membrane potential is given as:
where \(I_i^{net}(t)\) represents the synaptic current, which describes the sum of excitatory and inhibitory neurons.
where
Parameters: \(J_E = \frac 1 {\sqrt {pN_e}}, J_I = \frac 1 {\sqrt {pN_i}}\)
We can see from the dynamic that network is based on leaky Integrate-and-Fire neurons, and we can just use get_LIF from bpmodels.neurons to get this model.
[2]:
tau = 10.
V_rest = -52.
V_reset = -60.
V_th = -50.
Ib = 3.
[3]:
class LIF(bp.NeuGroup):
target_backend = 'numpy'
def __init__(self, size, **kwargs):
super(LIF, self).__init__(size, **kwargs)
self.V = bp.math.Variable(bp.math.zeros(size))
self.spike = bp.math.Variable(bp.math.zeros(size))
self.input = bp.math.Variable(bp.math.zeros(size))
self.integral = bp.odeint(self.derivative)
def derivative(self, V, t, Isyn):
return (-V + V_rest + Isyn) / tau
def update(self, _t, _dt):
for i in range(self.num):
V = self.integral(self.V[i], _t, self.input[i])
if V >= V_th:
self.spike[i] = 1.
V = V_reset
else:
self.spike[i] = 0.
self.V[i] = V
self.input[i] = Ib
The function of \(I_i^{net}(t)\) is actually a synase with single exponential decay, we can also get it by using get_exponential.
[4]:
tau_decay = 2.
[5]:
class Syn(bp.TwoEndConn):
target_backend = 'numpy'
def __init__(self, pre, post, conn, g_max, **kwargs):
super(Syn, self).__init__(pre, post, conn=conn, **kwargs)
# parameters
self.g_max = g_max
# connection
self.pre2post = self.conn.requires('pre2post')
# variables
self.s = bp.math.Variable(bp.math.zeros(post.num))
self.integral = bp.odeint(self.derivative)
def derivative(self, s, t):
return - s / tau_decay
def update(self, _t, _dt):
self.s[:] = self.integral(self.s, _t)
for pre_i, spike in enumerate(self.pre.spike):
if spike:
for post_i in self.pre2post[pre_i]:
self.s[post_i] += 1.
self.post.input += self.g_max * self.s
Network
Let’s create a neuron group with \(N_E\) excitatory neurons and \(N_I\) inbibitory neurons. Use conn=bp.connect.FixedProb(p) to implement the random and sparse connections.
[6]:
num_exc = 500
num_inh = 500
prob = 0.1
JE = 1 / bp.math.sqrt(prob * num_exc)
JI = 1 / bp.math.sqrt(prob * num_inh)
E = LIF(num_exc, monitors=['spike'])
E.V[:] = bp.math.random.random(num_exc) * (V_th - V_rest) + V_rest
I = LIF(num_inh, monitors=['spike'])
I.V[:] = bp.math.random.random(num_inh) * (V_th - V_rest) + V_rest
E2E = Syn(E, E, conn=bp.connect.FixedProb(prob=prob), g_max=JE)
E2I = Syn(E, I, conn=bp.connect.FixedProb(prob=prob), g_max=JE)
I2E = Syn(I, E, conn=bp.connect.FixedProb(prob=prob), g_max=-JI)
I2I = Syn(I, I, conn=bp.connect.FixedProb(prob=prob), g_max=-JI)
net = bp.Network(E, I, E2E, E2I, I2E, I2I)
net = bp.math.jit(net)
net.run(duration=1000., report=0.1)
Compilation used 3.8000 s.
Start running ...
Run 10.0% used 0.024 s.
Run 20.0% used 0.048 s.
Run 30.0% used 0.072 s.
Run 40.0% used 0.158 s.
Run 50.0% used 0.182 s.
Run 60.0% used 0.208 s.
Run 70.0% used 0.238 s.
Run 80.0% used 0.279 s.
Run 90.0% used 0.304 s.
Run 100.0% used 0.328 s.
Simulation is done in 0.328 s.
[6]:
0.3284895420074463
Visualization
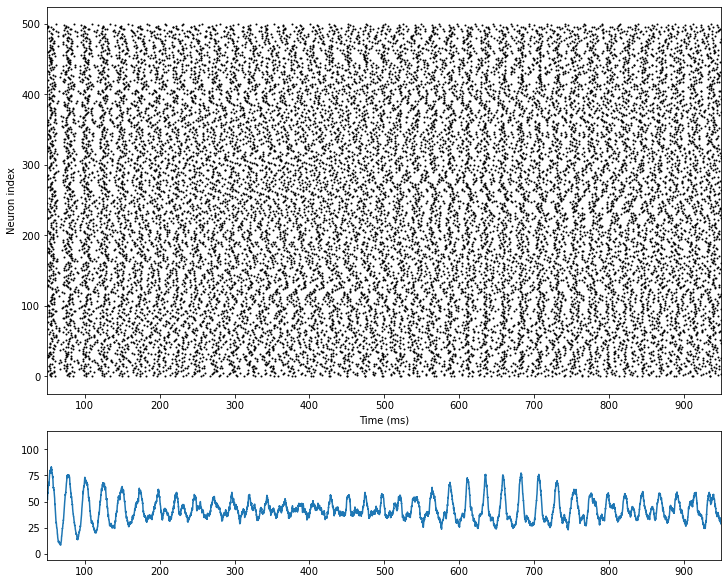
[7]:
import matplotlib.pyplot as plt
fig, gs = bp.visualize.get_figure(4, 1, 2, 10)
fig.add_subplot(gs[:3, 0])
bp.visualize.raster_plot(E.mon.ts, E.mon.spike, xlim=(50, 950))
fig.add_subplot(gs[3, 0])
rates = bp.measure.firing_rate(E.mon.spike, 5.)
plt.plot(E.mon.ts, rates)
plt.xlim(50, 950)
plt.show()
Reference
[1] Van Vreeswijk, Carl, and Haim Sompolinsky. “Chaos in neuronal networks with balanced excitatory and inhibitory activity.” Science 274.5293 (1996): 1724-1726.