Hindmarsh-Rose model bursting analysis
[1]:
import brainpy as bp
import brainpy.math as bm
The Hindmarsh-Rose model describes bursting dynamics in neurons.
“x” models the membrane potential of a bursting cell, “y” models the main currents into and out of the cell, and “z” models an adaptive (calcium-like) current.
[2]:
class HR(bp.NeuGroup):
@staticmethod
def derivative(V, y, z, t, a, b, c, d, r, s, x_r, Isyn):
dV = y - a * V ** 3 + b * V * V - z + Isyn
dy = c - d * V * V - y
dz = r * (s * (V - x_r) - z)
return dV, dy, dz
def __init__(self, size, a=1., b=3., c=1., d=5., s=4., x_r=-1.6,
r=0.001, Vth=1.9, **kwargs):
super(HR, self).__init__(size=size, **kwargs)
# parameters
self.a = a
self.b = b
self.c = c
self.d = d
self.s = s
self.x_r = x_r
self.r = r
self.Vth = Vth
# variables
self.V = bm.Variable(-1.6 * bm.ones(self.num))
self.y = bm.Variable(-10 * bm.ones(self.num))
self.z = bm.Variable(bm.zeros(self.num))
self.spike = bm.Variable(bm.zeros(self.num, dtype=bool))
self.input = bm.Variable(bm.zeros(self.num))
self.integral = bp.odeint(f=self.derivative)
def update(self, _t, _dt):
V, y, z = self.integral(self.V, self.y, self.z, _t,
self.a, self.b, self.c, self.d,
self.r, self.s, self.x_r, self.input)
self.spike[:] = (V >= self.Vth) * (self.V < self.Vth)
self.V[:] = V
self.y[:] = y
self.z[:] = z
self.input[:] = 0.
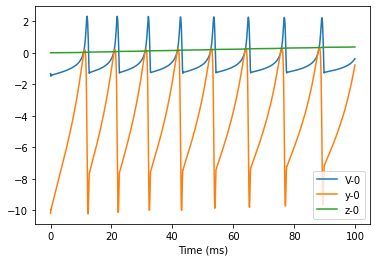
Simulation
[3]:
group = HR(1, monitors=['V', 'y', 'z'])
group.run(100., inputs=('input', 1.))
bp.visualize.line_plot(group.mon.ts, group.mon.V, legend='V', )
bp.visualize.line_plot(group.mon.ts, group.mon.y, legend='y', )
bp.visualize.line_plot(group.mon.ts, group.mon.z, legend='z', show=True)
WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
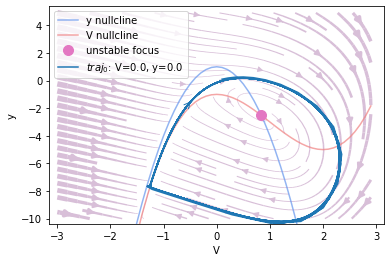
Phase plane analysis
[4]:
analyzer = bp.symbolic.PhasePlane(
group.integral,
target_vars={'V': [-3, 3], 'y': [-10., 5.]},
fixed_vars={'z': 0.},
pars_update=dict(a=1., b=3., c=1., d=5., s=4., x_r=-1.6, r=0.001, Isyn=1.)
)
analyzer.plot_nullcline()
analyzer.plot_vector_field()
analyzer.plot_fixed_point()
analyzer.plot_trajectory([{'V': 0., 'y': 0.}],
duration=100.,
show=True)
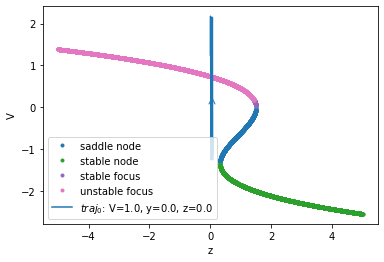
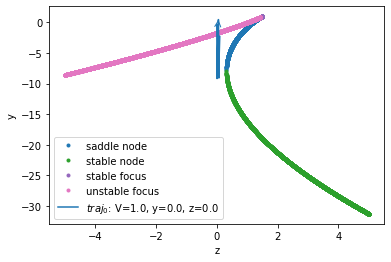
Bifurcation Diagram
[5]:
analyzer = bp.symbolic.FastSlowBifurcation(
group.integral,
fast_vars={'V': [-3, 3], 'y': [-10., 5.]},
slow_vars={'z': [-5., 5.]},
pars_update=dict(a=1., b=3., c=1., d=5., s=4., x_r=-1.6, r=0.001, Isyn=.5),
numerical_resolution=0.001
)
analyzer.plot_bifurcation()
analyzer.plot_trajectory([{'V': 1., 'y': 0., 'z': 0.0}],
duration=30.,
show=True)
References:
James L Hindmarsh and RM Rose. A model of neuronal bursting using three coupled first order differential equations. Proceedings of the Royal society of London. Series B. Biological sciences, 221(1222):87–102, 1984.