Train RNN to Solve Parametric Working Memory
[2]:
import brainpy as bp
bp.set_platform('cpu')
bp.math.use_backend('jax')
import brainpy.math.jax as bm
import brainpy.simulation.layers as layers
[3]:
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
[1]:
# We will import the task from the neurogym library.
# Please install neurogym:
#
# https://github.com/neurogym/neurogym
import neurogym as ngym
[4]:
# Environment
task = 'DelayComparison-v0'
timing = {'delay': ('choice', [200, 400, 800, 1600, 3200]),
'response': ('constant', 500)}
kwargs = {'dt': 100, 'timing': timing}
seq_len = 100
# Make supervised dataset
dataset = ngym.Dataset(task,
env_kwargs=kwargs,
batch_size=16,
seq_len=seq_len)
# A sample environment from dataset
env = dataset.env
# Visualize the environment with 2 sample trials
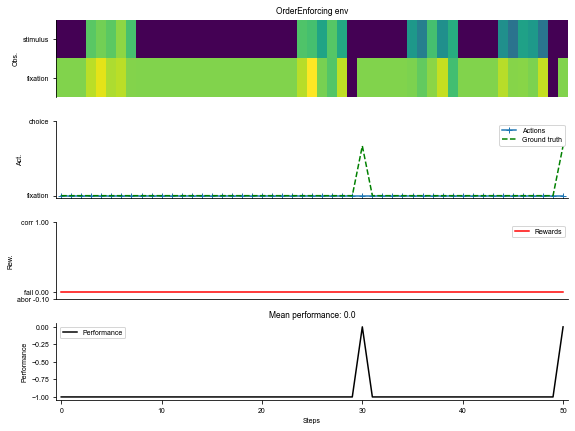
_ = ngym.utils.plot_env(env, num_trials=2, def_act=0, fig_kwargs={'figsize': (8, 6)})
plt.show()
[5]:
input_size = env.observation_space.shape[0]
output_size = env.action_space.n
batch_size = dataset.batch_size
[6]:
class RNN(layers.Module):
def __init__(self, num_input, num_hidden, num_output, num_batch, dt=None, seed=None,
w_ir=bp.init.KaimingNormal(scale=1.),
w_rr=bp.init.KaimingNormal(scale=1.),
w_ro=bp.init.KaimingNormal(scale=1.)):
super(RNN, self).__init__()
# parameters
self.tau = 100
self.num_batch = num_batch
self.num_input = num_input
self.num_hidden = num_hidden
self.num_output = num_output
if dt is None:
self.alpha = 1
else:
self.alpha = dt / self.tau
self.rng = bm.random.RandomState(seed=seed)
# input weight
self.w_ir = self.get_param(w_ir, (num_input, num_hidden))
# recurrent weight
bound = 1 / num_hidden ** 0.5
self.w_rr = self.get_param(w_rr, (num_hidden, num_hidden))
self.b_rr = bm.TrainVar(self.rng.uniform(-bound, bound, num_hidden))
# readout weight
self.w_ro = self.get_param(w_ro, (num_hidden, num_output))
self.b_ro = bm.TrainVar(self.rng.uniform(-bound, bound, num_output))
# variables
self.h = bm.Variable(bm.zeros((num_batch, num_hidden)))
self.o = bm.Variable(bm.zeros((num_batch, num_output)))
def cell(self, x, h):
ins = x @ self.w_ir + h @ self.w_rr + self.b_rr
state = h * (1 - self.alpha) + ins * self.alpha
return bm.relu(state)
def readout(self, h):
return h @ self.w_ro + self.b_ro
def make_update(self, h: bm.JaxArray, o: bm.JaxArray):
def f(x):
h.value = self.cell(x, h.value)
o.value = self.readout(h.value)
return f
def predict(self, xs):
self.h[:] = 0.
f = bm.make_loop(self.make_update(self.h, self.o),
dyn_vars=self.vars(),
out_vars=[self.h, self.o])
return f(xs)
def loss(self, xs, ys):
hs, os = self.predict(xs)
os = os.reshape((-1, os.shape[-1]))
loss = bm.losses.cross_entropy_loss(os, ys.flatten())
return loss, os
[7]:
# Instantiate the network and print information
hidden_size = 64
net = RNN(num_input=input_size,
num_hidden=hidden_size,
num_output=output_size,
num_batch=batch_size,
dt=env.dt)
[8]:
predict = bm.jit(net.predict, dyn_vars=net.vars())
[9]:
# Adam optimizer
opt = bm.optimizers.Adam(lr=0.001, train_vars=net.train_vars().unique())
[10]:
# gradient function
grad_f = bm.grad(net.loss,
dyn_vars=net.vars(),
grad_vars=net.train_vars().unique(),
return_value=True,
has_aux=True)
[11]:
@bm.jit
@bm.function(nodes=(net, opt))
def train(xs, ys):
grads, (loss, os) = grad_f(xs, ys)
opt.update(grads)
return loss, os
[12]:
running_acc = 0
running_loss = 0
for i in range(2000):
inputs, labels_np = dataset()
inputs = bm.asarray(inputs)
labels = bm.asarray(labels_np)
loss, outputs = train(inputs, labels)
running_loss += loss
# Compute performance
output_np = np.argmax(outputs.numpy(), axis=-1).flatten()
labels_np = labels_np.flatten()
ind = labels_np > 0 # Only analyze time points when target is not fixation
running_acc += np.mean(labels_np[ind] == output_np[ind])
if i % 100 == 99:
running_loss /= 100
running_acc /= 100
print('Step {}, Loss {:0.4f}, Acc {:0.3f}'.format(i + 1, running_loss, running_acc))
running_loss = 0
running_acc = 0
Step 100, Loss 0.1960, Acc 0.134
Step 200, Loss 0.0327, Acc 0.724
Step 300, Loss 0.0203, Acc 0.811
Step 400, Loss 0.0155, Acc 0.848
Step 500, Loss 0.0126, Acc 0.881
Step 600, Loss 0.0107, Acc 0.901
Step 700, Loss 0.0096, Acc 0.904
Step 800, Loss 0.0093, Acc 0.907
Step 900, Loss 0.0081, Acc 0.912
Step 1000, Loss 0.0081, Acc 0.913
Step 1100, Loss 0.0076, Acc 0.924
Step 1200, Loss 0.0071, Acc 0.927
Step 1300, Loss 0.0069, Acc 0.927
Step 1400, Loss 0.0083, Acc 0.913
Step 1500, Loss 0.0081, Acc 0.911
Step 1600, Loss 0.0071, Acc 0.923
Step 1700, Loss 0.0076, Acc 0.920
Step 1800, Loss 0.0065, Acc 0.927
Step 1900, Loss 0.0071, Acc 0.923
Step 2000, Loss 0.0066, Acc 0.927
[13]:
def run(num_trial=1):
env.reset(no_step=True)
perf = 0
activity_dict = {}
trial_infos = {}
for i in range(num_trial):
env.new_trial()
ob, gt = env.ob, env.gt
inputs = bm.asarray(ob[:, np.newaxis, :])
rnn_activity, action_pred = predict(inputs)
rnn_activity = rnn_activity.numpy()[:, 0, :]
activity_dict[i] = rnn_activity
trial_infos[i] = env.trial
# Concatenate activity for PCA
activity = np.concatenate(list(activity_dict[i] for i in range(num_trial)), axis=0)
print('Shape of the neural activity: (Time points, Neurons): ', activity.shape)
# Print trial informations
for i in range(5):
if i >= num_trial: break
print('Trial ', i, trial_infos[i])
pca = PCA(n_components=2)
pca.fit(activity)
# print('Shape of the projected activity: (Time points, PCs): ', activity_pc.shape)
fig, (ax1, ax2) = plt.subplots(1, 2, sharey=True, sharex=True, figsize=(6, 3))
for i in range(num_trial):
activity_pc = pca.transform(activity_dict[i])
trial = trial_infos[i]
color = 'red' if trial['ground_truth'] == 0 else 'blue'
_ = ax1.plot(activity_pc[:, 0], activity_pc[:, 1], 'o-', color=color)
if i < 3:
_ = ax2.plot(activity_pc[:, 0], activity_pc[:, 1], 'o-', color=color)
ax1.set_xlabel('PC 1')
ax1.set_ylabel('PC 2')
plt.show()
[14]:
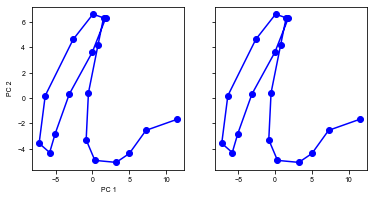
run(num_trial=1)
Shape of the neural activity: (Time points, Neurons): (18, 64)
Trial 0 {'ground_truth': 1, 'vpair': (30, 22), 'v1': 30, 'v2': 22}
[15]:
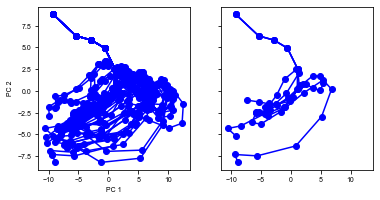
run(num_trial=20)
Shape of the neural activity: (Time points, Neurons): (548, 64)
Trial 0 {'ground_truth': 2, 'vpair': (34, 26), 'v1': 26, 'v2': 34}
Trial 1 {'ground_truth': 2, 'vpair': (30, 22), 'v1': 22, 'v2': 30}
Trial 2 {'ground_truth': 1, 'vpair': (18, 10), 'v1': 18, 'v2': 10}
Trial 3 {'ground_truth': 1, 'vpair': (30, 22), 'v1': 30, 'v2': 22}
Trial 4 {'ground_truth': 1, 'vpair': (34, 26), 'v1': 34, 'v2': 26}
[16]:
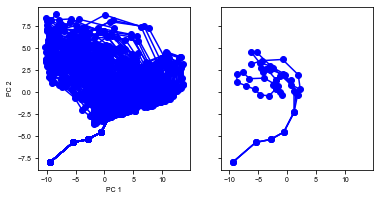
run(num_trial=100)
Shape of the neural activity: (Time points, Neurons): (2862, 64)
Trial 0 {'ground_truth': 1, 'vpair': (26, 18), 'v1': 26, 'v2': 18}
Trial 1 {'ground_truth': 1, 'vpair': (22, 14), 'v1': 22, 'v2': 14}
Trial 2 {'ground_truth': 2, 'vpair': (18, 10), 'v1': 10, 'v2': 18}
Trial 3 {'ground_truth': 2, 'vpair': (26, 18), 'v1': 18, 'v2': 26}
Trial 4 {'ground_truth': 2, 'vpair': (34, 26), 'v1': 26, 'v2': 34}