(Mi, et. al., 2017) STP for Working Memory Capacity
Implementation of the paper:
Mi, Yuanyuan, Mikhail Katkov, and Misha Tsodyks. “Synaptic correlates of working memory capacity.” Neuron 93.2 (2017): 323-330.
[1]:
import matplotlib.colors as mcolors
import matplotlib.pyplot as plt
[2]:
import brainpy as bp
import brainpy.math as bm
bm.enable_x64(True)
bm.set_platform('cpu')
[3]:
alpha = 1.5
J_EE = 8. # the connection strength in each excitatory neural clusters
J_IE = 1.75 # Synaptic efficacy E → I
J_EI = 1.1 # Synaptic efficacy I → E
tau_f = 1.5 # time constant of STF [s]
tau_d = .3 # time constant of STD [s]
U = 0.3 # minimum STF value
tau = 0.008 # time constant of firing rate of the excitatory neurons [s]
tau_I = tau # time constant of firing rate of the inhibitory neurons
Ib = 8. # background input and external input
Iinh = 0. # the background input of inhibtory neuron
cluster_num = 16 # the number of the clusters
[4]:
# the parameters of external input
stimulus_num = 5
Iext_train = 225 # the strength of the external input
Ts_interval = 0.070 # the time interval between the consequent external input [s]
Ts_duration = 0.030 # the time duration of the external input [s]
duration = 2.500 # [s]
[5]:
# the excitatory cluster model and the inhibitory pool model
class WorkingMemoryModel(bp.DynamicalSystem):
def __init__(self, **kwargs):
super(WorkingMemoryModel, self).__init__(**kwargs)
# variables
self.u = bm.Variable(bm.ones(cluster_num) * U)
self.x = bm.Variable(bm.ones(cluster_num))
self.h = bm.Variable(bm.zeros(cluster_num))
self.r = bm.Variable(self.log(self.h))
self.input = bm.Variable(bm.zeros(cluster_num))
self.inh_h = bm.Variable(bm.zeros(1))
self.inh_r = bm.Variable(self.log(self.inh_h))
def log(self, h):
# return alpha * bm.log(1. + bm.exp(h / alpha))
return alpha * bm.log1p(bm.exp(h / alpha))
def update(self, _t, _dt):
uxr = self.u * self.x * self.r
du = (U - self.u) / tau_f + U * (1 - self.u) * self.r
dx = (1 - self.x) / tau_d - uxr
dh = (-self.h + J_EE * uxr - J_EI * self.inh_r + self.input + Ib) / tau
dhi = (-self.inh_h + J_IE * bm.sum(self.r) + Iinh) / tau_I
self.u += du * _dt
self.x += dx * _dt
self.h += dh * _dt
self.inh_h += dhi * _dt
self.r[:] = self.log(self.h)
self.inh_r[:] = self.log(self.inh_h)
self.input[:] = 0.
[6]:
dt = 0.0001 # [s]
# the external input
I_inputs = bm.zeros((int(duration / dt), cluster_num))
for i in range(stimulus_num):
t_start = (Ts_interval + Ts_duration) * i + Ts_interval
t_end = t_start + Ts_duration
idx_start, idx_end = int(t_start / dt), int(t_end / dt)
I_inputs[idx_start: idx_end, i] = Iext_train
[7]:
# running
runner = bp.StructRunner(WorkingMemoryModel(),
inputs=['input', I_inputs, 'iter'],
monitors=['u', 'x', 'r', 'h'],
dt=dt)
runner(duration)
[7]:
0.2612760066986084
[8]:
# visualization
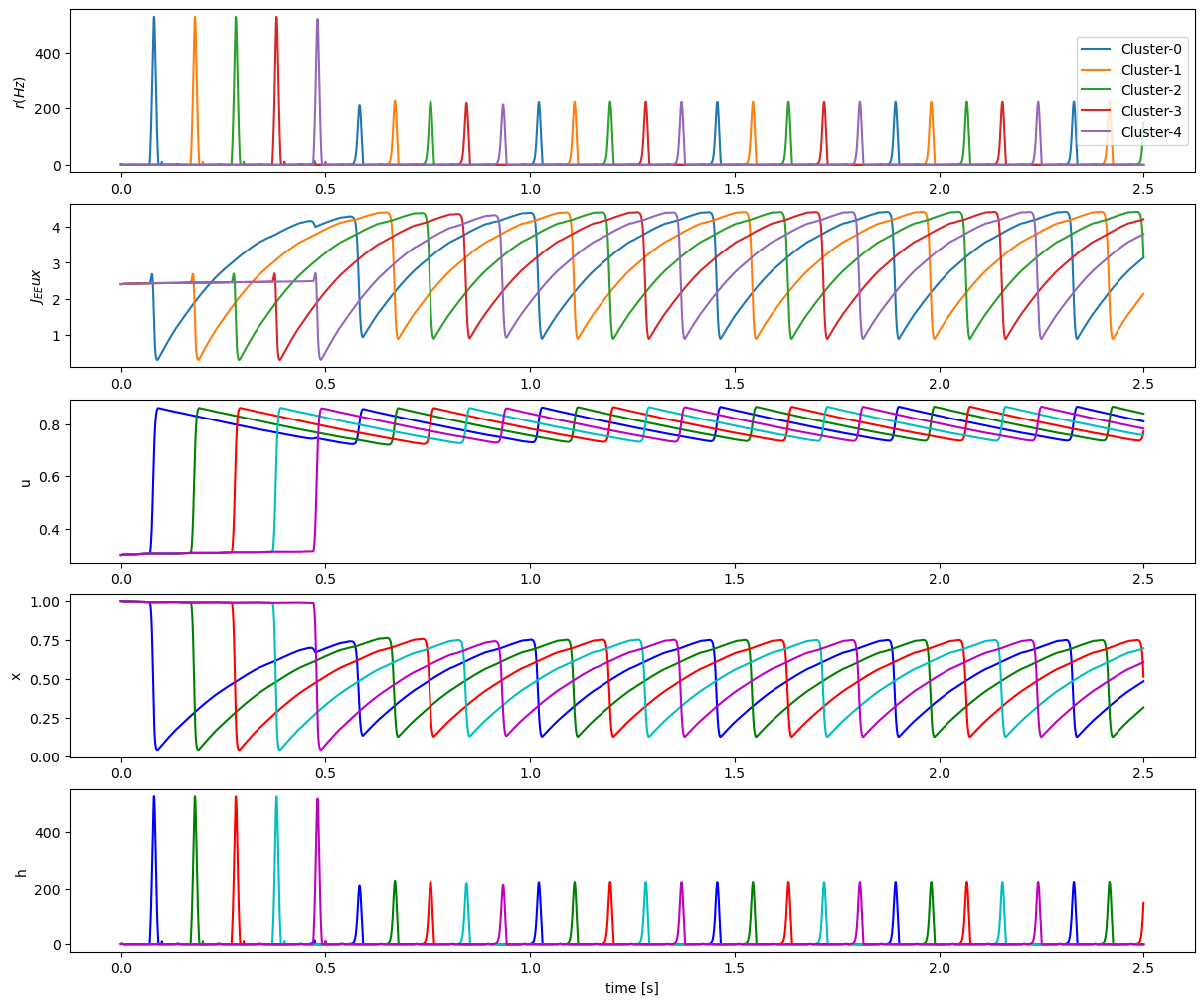
runner.mon.numpy()
colors = list(dict(mcolors.BASE_COLORS, **mcolors.CSS4_COLORS).keys())
fig, gs = bp.visualize.get_figure(5, 1, 2, 12)
fig.add_subplot(gs[0, 0])
for i in range(stimulus_num):
plt.plot(runner.mon.ts, runner.mon.r[:, i], label='Cluster-{}'.format(i))
plt.ylabel("$r (Hz)$")
plt.legend(loc='right')
fig.add_subplot(gs[1, 0])
hist_Jux = J_EE * runner.mon.u * runner.mon.x
for i in range(stimulus_num):
plt.plot(runner.mon.ts, hist_Jux[:, i])
plt.ylabel("$J_{EE}ux$")
fig.add_subplot(gs[2, 0])
for i in range(stimulus_num):
plt.plot(runner.mon.ts, runner.mon.u[:, i], colors[i])
plt.ylabel('u')
fig.add_subplot(gs[3, 0])
for i in range(stimulus_num):
plt.plot(runner.mon.ts, runner.mon.x[:, i], colors[i])
plt.ylabel('x')
fig.add_subplot(gs[4, 0])
for i in range(stimulus_num):
plt.plot(runner.mon.ts, runner.mon.r[:, i], colors[i])
plt.ylabel('h')
plt.xlabel('time [s]')
plt.show()