Gap junction-coupled FitzHugh-Nagumo Model
[1]:
import brainpy as bp
import brainpy.math as bm
# bp.math.set_platform('cpu')
bp.math.enable_x64()
[2]:
class GJCoupledFHN(bp.DynamicalSystem):
def __init__(self, num=2, method='exp_auto'):
super(GJCoupledFHN, self).__init__()
# parameters
self.num = num
self.a = 0.7
self.b = 0.8
self.tau = 12.5
self.gjw = 0.0001
# variables
self.V = bm.Variable(bm.random.uniform(-2, 2, num))
self.w = bm.Variable(bm.random.uniform(-2, 2, num))
self.Iext = bm.Variable(bm.zeros(num))
# functions
self.int_V = bp.odeint(self.dV, method=method)
self.int_w = bp.odeint(self.dw, method=method)
def dV(self, V, t, w, Iext=0.):
gj = (V.reshape((-1, 1)) - V).sum(axis=0) * self.gjw
dV = V - V * V * V / 3 - w + Iext + gj
return dV
def dw(self, w, t, V):
dw = (V + self.a - self.b * w) / self.tau
return dw
def update(self, tdi):
self.V.value = self.int_V(self.V, tdi.t, self.w, self.Iext, tdi.dt)
self.w.value = self.int_w(self.w, tdi.t, self.V, tdi.dt)
[3]:
def analyze_net(num=2, gjw=0.01, Iext=bm.asarray([0., 0.6])):
assert isinstance(Iext, (int, float)) or (len(Iext) == num)
model = GJCoupledFHN(num)
model.gjw = gjw
model.Iext[:] = Iext
# simulation
runner = bp.DSRunner(model, monitors=['V'])
runner.run(300.)
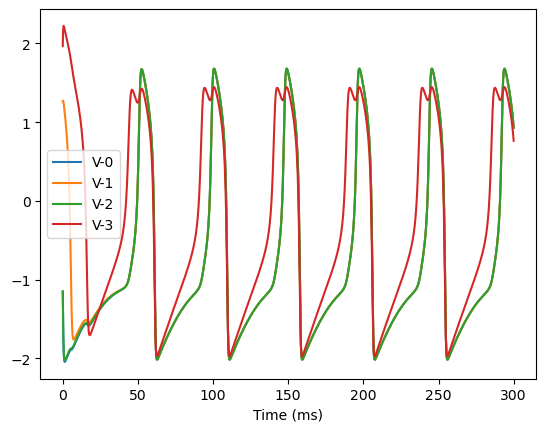
bp.visualize.line_plot(runner.mon.ts, runner.mon.V, legend='V',
plot_ids=list(range(model.num)), show=True)
# analysis
finder = bp.analysis.SlowPointFinder(f_cell=model,
target_vars={'V': model.V, 'w': model.w})
finder.find_fps_with_gd_method(
candidates={'V': bm.random.normal(0., 2., (1000, model.num)),
'w': bm.random.normal(0., 2., (1000, model.num))},
tolerance=1e-5,
num_batch=200,
optimizer=bp.optim.Adam(lr=bp.optim.ExponentialDecay(0.05, 1, 0.9999)),
)
finder.filter_loss(1e-8)
finder.keep_unique()
print('fixed_points: ', finder.fixed_points)
print('losses:', finder.losses)
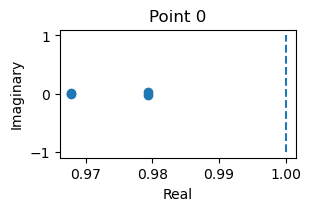
_ = finder.compute_jacobians(finder.fixed_points, plot=True)
WARNING:jax._src.lib.xla_bridge:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
4D system
[4]:
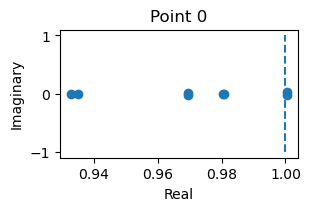
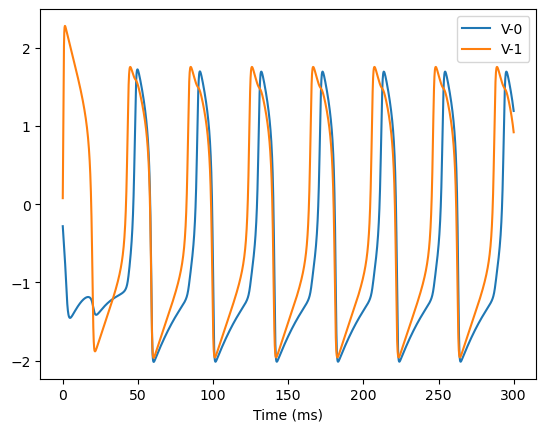
analyze_net(num=2, gjw=0.1, Iext=bm.asarray([0., 0.6]))
Optimizing with Adam(lr=ExponentialDecay(0.05, decay_steps=1, decay_rate=0.9999), last_call=-1), beta1=0.9, beta2=0.999, eps=1e-08) to find fixed points:
Batches 1-200 in 0.47 sec, Training loss 0.0002525318
Batches 201-400 in 1.82 sec, Training loss 0.0002021251
Batches 401-600 in 1.06 sec, Training loss 0.0001682558
Batches 601-800 in 0.64 sec, Training loss 0.0001426594
Batches 801-1000 in 0.88 sec, Training loss 0.0001222235
Batches 1001-1200 in 1.86 sec, Training loss 0.0001054805
Batches 1201-1400 in 0.51 sec, Training loss 0.0000914966
Batches 1401-1600 in 0.60 sec, Training loss 0.0000796636
Batches 1601-1800 in 0.75 sec, Training loss 0.0000695541
Batches 1801-2000 in 2.01 sec, Training loss 0.0000608892
Batches 2001-2200 in 0.63 sec, Training loss 0.0000534558
Batches 2201-2400 in 0.63 sec, Training loss 0.0000470366
Batches 2401-2600 in 1.92 sec, Training loss 0.0000414673
Batches 2601-2800 in 0.86 sec, Training loss 0.0000366226
Batches 2801-3000 in 0.44 sec, Training loss 0.0000323996
Batches 3001-3200 in 0.47 sec, Training loss 0.0000287081
Batches 3201-3400 in 0.65 sec, Training loss 0.0000254832
Batches 3401-3600 in 0.53 sec, Training loss 0.0000226843
Batches 3601-3800 in 0.80 sec, Training loss 0.0000202542
Batches 3801-4000 in 1.89 sec, Training loss 0.0000181205
Batches 4001-4200 in 0.64 sec, Training loss 0.0000162401
Batches 4201-4400 in 0.52 sec, Training loss 0.0000145707
Batches 4401-4600 in 2.00 sec, Training loss 0.0000130935
Batches 4601-4800 in 0.67 sec, Training loss 0.0000118004
Batches 4801-5000 in 0.59 sec, Training loss 0.0000106516
Batches 5001-5200 in 0.86 sec, Training loss 0.0000096305
Stop optimization as mean training loss 0.0000096305 is below tolerance 0.0000100000.
Excluding fixed points with squared speed above tolerance 1e-08:
Kept 306/1000 fixed points with tolerance under 1e-08.
Excluding non-unique fixed points:
Kept 1/306 unique fixed points with uniqueness tolerance 0.025.
fixed_points: {'V': array([[-1.1731516 , -0.73801606]]), 'w': array([[-0.59144118, -0.04753834]])}
losses: [6.8918834e-15]
[5]:
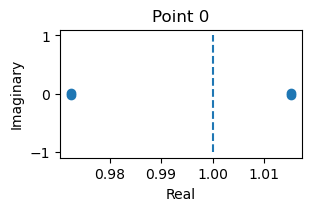
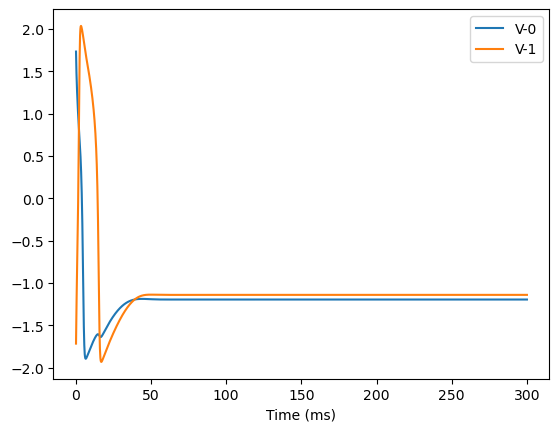
analyze_net(num=2, gjw=0.1, Iext=bm.asarray([0., 0.1]))
Optimizing with Adam(lr=ExponentialDecay(0.05, decay_steps=1, decay_rate=0.9999), last_call=-1), beta1=0.9, beta2=0.999, eps=1e-08) to find fixed points:
Batches 1-200 in 1.39 sec, Training loss 0.0002524889
Batches 201-400 in 0.47 sec, Training loss 0.0002039485
Batches 401-600 in 0.39 sec, Training loss 0.0001722068
Batches 601-800 in 0.49 sec, Training loss 0.0001481642
Batches 801-1000 in 0.67 sec, Training loss 0.0001288039
Batches 1001-1200 in 0.69 sec, Training loss 0.0001127157
Batches 1201-1400 in 2.42 sec, Training loss 0.0000990425
Batches 1401-1600 in 0.71 sec, Training loss 0.0000872609
Batches 1601-1800 in 0.45 sec, Training loss 0.0000770819
Batches 1801-2000 in 1.56 sec, Training loss 0.0000682273
Batches 2001-2200 in 1.07 sec, Training loss 0.0000605319
Batches 2201-2400 in 0.59 sec, Training loss 0.0000538263
Batches 2401-2600 in 0.38 sec, Training loss 0.0000479664
Batches 2601-2800 in 1.37 sec, Training loss 0.0000428171
Batches 2801-3000 in 0.77 sec, Training loss 0.0000382962
Batches 3001-3200 in 0.53 sec, Training loss 0.0000343242
Batches 3201-3400 in 0.39 sec, Training loss 0.0000307939
Batches 3401-3600 in 1.54 sec, Training loss 0.0000276377
Batches 3601-3800 in 0.71 sec, Training loss 0.0000248339
Batches 3801-4000 in 0.41 sec, Training loss 0.0000223625
Batches 4001-4200 in 0.46 sec, Training loss 0.0000202066
Batches 4201-4400 in 0.58 sec, Training loss 0.0000182917
Batches 4401-4600 in 0.47 sec, Training loss 0.0000165570
Batches 4601-4800 in 1.13 sec, Training loss 0.0000150186
Batches 4801-5000 in 0.96 sec, Training loss 0.0000136606
Batches 5001-5200 in 0.50 sec, Training loss 0.0000124335
Batches 5201-5400 in 0.35 sec, Training loss 0.0000113094
Batches 5401-5600 in 1.40 sec, Training loss 0.0000103006
Batches 5601-5800 in 0.83 sec, Training loss 0.0000093835
Stop optimization as mean training loss 0.0000093835 is below tolerance 0.0000100000.
Excluding fixed points with squared speed above tolerance 1e-08:
Kept 485/1000 fixed points with tolerance under 1e-08.
Excluding non-unique fixed points:
Kept 1/485 unique fixed points with uniqueness tolerance 0.025.
fixed_points: {'V': array([[-1.19613916, -1.14106972]]), 'w': array([[-0.62017396, -0.55133715]])}
losses: [2.01213341e-24]
8D system
[6]:
analyze_net(num=4, gjw=0.1, Iext=bm.asarray([0., 0., 0., 0.6]))
Optimizing with Adam(lr=ExponentialDecay(0.05, decay_steps=1, decay_rate=0.9999), last_call=-1), beta1=0.9, beta2=0.999, eps=1e-08) to find fixed points:
Batches 1-200 in 0.58 sec, Training loss 0.0002563626
Batches 201-400 in 0.63 sec, Training loss 0.0002068401
Batches 401-600 in 1.46 sec, Training loss 0.0001746516
Batches 601-800 in 0.53 sec, Training loss 0.0001505581
Batches 801-1000 in 0.34 sec, Training loss 0.0001312724
Batches 1001-1200 in 0.39 sec, Training loss 0.0001152650
Batches 1201-1400 in 0.57 sec, Training loss 0.0001017105
Batches 1401-1600 in 0.36 sec, Training loss 0.0000900637
Batches 1601-1800 in 1.23 sec, Training loss 0.0000799450
Batches 1801-2000 in 0.90 sec, Training loss 0.0000710771
Batches 2001-2200 in 0.58 sec, Training loss 0.0000632785
Batches 2201-2400 in 0.39 sec, Training loss 0.0000564160
Batches 2401-2600 in 1.61 sec, Training loss 0.0000503661
Batches 2601-2800 in 0.55 sec, Training loss 0.0000449990
Batches 2801-3000 in 0.51 sec, Training loss 0.0000402453
Batches 3001-3200 in 0.72 sec, Training loss 0.0000360206
Batches 3201-3400 in 1.17 sec, Training loss 0.0000322757
Batches 3401-3600 in 0.47 sec, Training loss 0.0000289431
Batches 3601-3800 in 0.44 sec, Training loss 0.0000259914
Batches 3801-4000 in 0.49 sec, Training loss 0.0000233745
Batches 4001-4200 in 1.22 sec, Training loss 0.0000210534
Batches 4201-4400 in 0.56 sec, Training loss 0.0000189919
Batches 4401-4600 in 0.30 sec, Training loss 0.0000171629
Batches 4601-4800 in 0.33 sec, Training loss 0.0000155418
Batches 4801-5000 in 0.46 sec, Training loss 0.0000141007
Batches 5001-5200 in 0.32 sec, Training loss 0.0000128158
Batches 5201-5400 in 0.51 sec, Training loss 0.0000116671
Batches 5401-5600 in 1.17 sec, Training loss 0.0000106446
Batches 5601-5800 in 0.57 sec, Training loss 0.0000097291
Stop optimization as mean training loss 0.0000097291 is below tolerance 0.0000100000.
Excluding fixed points with squared speed above tolerance 1e-08:
Kept 266/1000 fixed points with tolerance under 1e-08.
Excluding non-unique fixed points:
Kept 1/266 unique fixed points with uniqueness tolerance 0.025.
fixed_points: {'V': array([[-1.17905465, -1.19006061, -1.17904012, -0.81757202]]), 'w': array([[-0.59764936, -0.58889147, -0.59766085, -0.14496394]])}
losses: [5.78896779e-09]