(Joglekar, et. al, 2018): Inter-areal Balanced Amplification Figure 5
Implementation of the figure 5 of:
Joglekar, Madhura R., et al. “Inter-areal balanced amplification enhances signal propagation in a large-scale circuit model of the primate cortex.” Neuron 98.1 (2018): 222-234.
[1]:
import brainpy as bp
import brainpy.math as bm
from brainpy.dyn import neurons
import matplotlib.pyplot as plt
import numpy as np
from jax import vmap
from scipy.io import loadmat
[2]:
# This model should be run on a GPU device
bm.set_platform('gpu')
[3]:
class MultiAreaNet(bp.Network):
def __init__(
self, hier, conn, delay_mat, muIE=0.0475, muEE=.0375, wII=.075,
wEE=.01, wIE=.075, wEI=.0375, extE=15.4, extI=14.0, alpha=4., seed=None,
):
super(MultiAreaNet, self).__init__()
# data
self.hier = hier
self.conn = conn
self.delay_mat = delay_mat
# parameters
self.muIE = muIE
self.muEE = muEE
self.wII = wII
self.wEE = wEE
self.wIE = wIE
self.wEI = wEI
self.extE = extE
self.extI = extI
self.alpha = alpha
num_area = hier.size
self.num_area = num_area
rng = bm.random.RandomState(seed)
# neuron models
self.E = neurons.LIF((num_area, 1600), V_th=-50., V_reset=-60.,
V_rest=-70., tau=20., tau_ref=2.,
noise=3. / bm.sqrt(20.),
V_initializer=bp.init.Uniform(-70., -50.),
method='euler',
keep_size=True)
self.I = neurons.LIF((num_area, 400), V_th=-50., V_reset=-60.,
V_rest=-70., tau=10., tau_ref=2., noise=3. / bm.sqrt(10.),
V_initializer=bp.init.Uniform(-70., -50.),
method='euler',
keep_size=True)
# delays
self.intra_delay_step = int(2. / bm.get_dt())
self.E_delay_steps = bm.asarray(delay_mat.T / bm.get_dt(), dtype=int)
bm.fill_diagonal(self.E_delay_steps, self.intra_delay_step)
self.Edelay = bm.LengthDelay(self.E.spike, delay_len=int(self.E_delay_steps.max()))
self.Idelay = bm.LengthDelay(self.I.spike, delay_len=self.intra_delay_step)
# synapse model
syn_fun = lambda pre_spike, weight, conn_mat: weight * (pre_spike @ conn_mat)
self.f_E_current = vmap(syn_fun)
self.f_I_current = vmap(syn_fun, in_axes=(0, None, 0))
# synapses from I
self.intra_I2E_conn = rng.random((num_area, 400, 1600)) < 0.1
self.intra_I2I_conn = rng.random((num_area, 400, 400)) < 0.1
self.intra_I2E_weight = -wEI
self.intra_I2I_weight = -wII
# synapses from E
self.E2E_conns = [rng.random((num_area, 1600, 1600)) < 0.1 for _ in range(num_area)]
self.E2I_conns = [rng.random((num_area, 1600, 400)) < 0.1 for _ in range(num_area)]
self.E2E_weights = (1 + alpha * hier) * muEE * conn.T # inter-area connections
bm.fill_diagonal(self.E2E_weights, (1 + alpha * hier) * wEE) # intra-area connections
self.E2I_weights = (1 + alpha * hier) * muIE * conn.T # inter-area connections
bm.fill_diagonal(self.E2I_weights, (1 + alpha * hier) * wIE) # intra-area connections
def update(self, tdi, v1_input):
self.E.input[0] += v1_input
self.E.input += self.extE
self.I.input += self.extI
E_not_ref = bm.logical_not(self.E.refractory)
I_not_ref = bm.logical_not(self.I.refractory)
# synapses from E
for i in range(self.num_area):
delayed_E_spikes = self.Edelay(self.E_delay_steps[i], i).astype(float)
current = self.f_E_current(delayed_E_spikes, self.E2E_weights[i], self.E2E_conns[i])
self.E.V += current * E_not_ref # E2E
current = self.f_E_current(delayed_E_spikes, self.E2I_weights[i], self.E2I_conns[i])
self.I.V += current * I_not_ref # E2I
# synapses from I
delayed_I_spikes = self.Idelay(self.intra_delay_step).astype(float)
current = self.f_I_current(delayed_I_spikes, self.intra_I2E_weight, self.intra_I2E_conn)
self.E.V += current * E_not_ref # I2E
current = self.f_I_current(delayed_I_spikes, self.intra_I2I_weight, self.intra_I2I_conn)
self.I.V += current * I_not_ref # I2I
# updates
self.Edelay.update(self.E.spike)
self.Idelay.update(self.I.spike)
self.E.update(tdi)
self.I.update(tdi)
[4]:
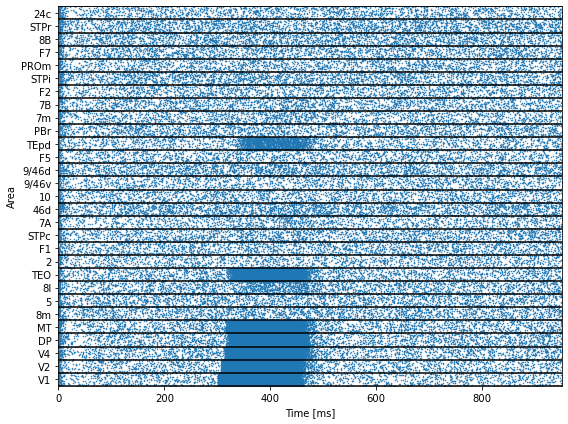
def raster_plot(xValues, yValues, duration):
ticks = np.round(np.arange(0, 29) + 0.5, 2)
areas = ['V1', 'V2', 'V4', 'DP', 'MT', '8m', '5', '8l', 'TEO', '2', 'F1',
'STPc', '7A', '46d', '10', '9/46v', '9/46d', 'F5', 'TEpd', 'PBr',
'7m', '7B', 'F2', 'STPi', 'PROm', 'F7', '8B', 'STPr', '24c']
N = len(ticks)
plt.figure(figsize=(8, 6))
plt.plot(xValues, yValues / (4 * 400), '.', markersize=1)
plt.plot([0, duration], np.arange(N + 1).repeat(2).reshape(-1, 2).T, 'k-')
plt.ylabel('Area')
plt.yticks(np.arange(N))
plt.xlabel('Time [ms]')
plt.ylim(0, N)
plt.yticks(ticks, areas)
plt.xlim(0, duration)
plt.tight_layout()
plt.show()
[5]:
# hierarchy values
hierVals = loadmat('Joglekar_2018_data/hierValspython.mat')
hierValsnew = hierVals['hierVals'].flatten()
hier = bm.asarray(hierValsnew / max(hierValsnew)) # hierarchy normalized.
# fraction of labeled neurons
flnMatp = loadmat('Joglekar_2018_data/efelenMatpython.mat')
conn = bm.asarray(flnMatp['flnMatpython'].squeeze()) # fln values..Cij is strength from j to i
# Distance
speed = 3.5 # axonal conduction velocity
distMatp = loadmat('Joglekar_2018_data/subgraphWiring29.mat')
distMat = distMatp['wiring'].squeeze() # distances between areas values..
delayMat = bm.asarray(distMat / speed)
[6]:
pars = dict(extE=14.2, extI=14.7, wII=.075, wEE=.01, wIE=.075, wEI=.0375, muEE=.0375, muIE=0.0475)
inps = dict(value=15, duration=150)
[7]:
inputs, length = bp.inputs.section_input(values=[0, inps['value'], 0.],
durations=[300., inps['duration'], 500],
return_length=True)
[8]:
net = MultiAreaNet(hier, conn, delayMat, **pars)
runner = bp.DSRunner(net, fun_monitors={'E.spike': lambda tdi: net.E.spike.flatten()})
runner.run(length, inputs=inputs)
[9]:
times, indices = np.where(runner.mon['E.spike'])
times = runner.mon.ts[times]
raster_plot(times, indices, length)