(Li, et. al, 2017): Unified Thalamus Oscillation Model
Implementation of the model:
Li, Guoshi, Craig S. Henriquez, and Flavio Fröhlich. “Unified thalamic model generates multiple distinct oscillations with state-dependent entrainment by stimulation.” PLoS computational biology 13.10 (2017): e1005797.
[1]:
from typing import Dict
import matplotlib.pyplot as plt
import numpy as np
import brainpy as bp
import brainpy.math as bm
from brainpy import channels, synapses, synouts, synplast
HTC neuron
[2]:
class HTC(bp.CondNeuGroup):
def __init__(self, size, gKL=0.01, V_initializer=bp.init.OneInit(-65.), ):
gL = 0.01 if size == 1 else bp.init.Uniform(0.0075, 0.0125)
IL = channels.IL(size, g_max=gL, E=-70)
IKL = channels.IKL(size, g_max=gKL)
INa = channels.INa_Ba2002(size, V_sh=-30)
IDR = channels.IKDR_Ba2002(size, V_sh=-30., phi=0.25)
Ih = channels.Ih_HM1992(size, g_max=0.01, E=-43)
ICaL = channels.ICaL_IS2008(size, g_max=0.5)
IAHP = channels.IAHP_De1994(size, g_max=0.3, E=-90.)
ICaN = channels.ICaN_IS2008(size, g_max=0.5)
ICaT = channels.ICaT_HM1992(size, g_max=2.1)
ICaHT = channels.ICaHT_HM1992(size, g_max=3.0)
Ca = channels.CalciumDetailed(size, C_rest=5e-5, tau=10., d=0.5, ICaL=ICaL,
IAHP=IAHP, ICaN=ICaN, ICaT=ICaT, ICaHT=ICaHT)
super(HTC, self).__init__(size, A=2.9e-4, V_initializer=V_initializer, V_th=20.,
IL=IL, IKL=IKL, INa=INa, IDR=IDR, Ih=Ih, Ca=Ca)
RTC neuron
[3]:
class RTC(bp.CondNeuGroup):
def __init__(self, size, gKL=0.01, V_initializer=bp.init.OneInit(-65.), ):
gL = 0.01 if size == 1 else bp.init.Uniform(0.0075, 0.0125)
IL = channels.IL(size, g_max=gL, E=-70)
IKL = channels.IKL(size, g_max=gKL)
INa = channels.INa_Ba2002(size, V_sh=-40)
IDR = channels.IKDR_Ba2002(size, V_sh=-40, phi=0.25)
Ih = channels.Ih_HM1992(size, g_max=0.01, E=-43)
ICaL = channels.ICaL_IS2008(size, g_max=0.3)
IAHP = channels.IAHP_De1994(size, g_max=0.1, E=-90.)
ICaN = channels.ICaN_IS2008(size, g_max=0.6)
ICaT = channels.ICaT_HM1992(size, g_max=2.1)
ICaHT = channels.ICaHT_HM1992(size, g_max=0.6)
Ca = channels.CalciumDetailed(size, C_rest=5e-5, tau=10., d=0.5, ICaL=ICaL,
IAHP=IAHP, ICaN=ICaN, ICaT=ICaT, ICaHT=ICaHT)
super(RTC, self).__init__(size, A=2.9e-4, V_initializer=V_initializer, V_th=20.,
IL=IL, IKL=IKL, INa=INa, IDR=IDR, Ih=Ih, Ca=Ca)
IN neuron
[4]:
class IN(bp.CondNeuGroup):
def __init__(self, size, gKL=0.01, V_initializer=bp.init.OneInit(-70.), ):
gL = 0.01 if size == 1 else bp.init.Uniform(0.0075, 0.0125)
IL = channels.IL(size, g_max=gL, E=-60)
IKL = channels.IKL(size, g_max=gKL)
INa = channels.INa_Ba2002(size, V_sh=-30)
IDR = channels.IKDR_Ba2002(size, V_sh=-30, phi=0.25)
Ih = channels.Ih_HM1992(size, g_max=0.05, E=-43)
IAHP = channels.IAHP_De1994(size, g_max=0.2, E=-90.)
ICaN = channels.ICaN_IS2008(size, g_max=0.1)
ICaHT = channels.ICaHT_HM1992(size, g_max=2.5)
Ca = channels.CalciumDetailed(size, C_rest=5e-5, tau=10., d=0.5,
IAHP=IAHP, ICaN=ICaN, ICaHT=ICaHT)
super(IN, self).__init__(size, A=1.7e-4, V_initializer=V_initializer, V_th=20.,
IL=IL, IKL=IKL, INa=INa, IDR=IDR, Ih=Ih, Ca=Ca)
TRN neuron
[5]:
class TRN(bp.CondNeuGroup):
def __init__(self, size, gKL=0.01, V_initializer=bp.init.OneInit(-70.), ):
gL = 0.01 if size == 1 else bp.init.Uniform(0.0075, 0.0125)
IL = channels.IL(size, g_max=gL, E=-60)
IKL = channels.IKL(size, g_max=gKL)
INa = channels.INa_Ba2002(size, V_sh=-40)
IDR = channels.IKDR_Ba2002(size, V_sh=-40)
IAHP = channels.IAHP_De1994(size, g_max=0.2, E=-90.)
ICaN = channels.ICaN_IS2008(size, g_max=0.2)
ICaT = channels.ICaT_HP1992(size, g_max=1.3)
Ca = channels.CalciumDetailed(size, C_rest=5e-5, tau=100., d=0.5,
IAHP=IAHP, ICaN=ICaN, ICaT=ICaT)
super(TRN, self).__init__(size, A=1.43e-4,
V_initializer=V_initializer, V_th=20.,
IL=IL, IKL=IKL, INa=INa, IDR=IDR, Ca=Ca)
Thalamus network
[6]:
class MgBlock(bp.SynOut):
def __init__(self, E=0.):
super(MgBlock, self).__init__()
self.E = E
def filter(self, g):
V = self.master.post.V.value
return g * (self.E - V) / (1 + bm.exp(-(V + 25) / 12.5))
[7]:
class Thalamus(bp.Network):
def __init__(
self,
g_input: Dict[str, float],
g_KL: Dict[str, float],
HTC_V_init=bp.init.OneInit(-65.),
RTC_V_init=bp.init.OneInit(-65.),
IN_V_init=bp.init.OneInit(-70.),
RE_V_init=bp.init.OneInit(-70.),
):
super(Thalamus, self).__init__()
# populations
self.HTC = HTC(size=(7, 7), gKL=g_KL['TC'], V_initializer=HTC_V_init)
self.RTC = RTC(size=(12, 12), gKL=g_KL['TC'], V_initializer=RTC_V_init)
self.RE = TRN(size=(10, 10), gKL=g_KL['RE'], V_initializer=IN_V_init)
self.IN = IN(size=(8, 8), gKL=g_KL['IN'], V_initializer=RE_V_init)
# noises
self.poisson_HTC = bp.neurons.PoissonGroup(self.HTC.size, freqs=100)
self.poisson_RTC = bp.neurons.PoissonGroup(self.RTC.size, freqs=100)
self.poisson_IN = bp.neurons.PoissonGroup(self.IN.size, freqs=100)
self.poisson_RE = bp.neurons.PoissonGroup(self.RE.size, freqs=100)
self.noise2HTC = synapses.Exponential(self.poisson_HTC, self.HTC, bp.conn.One2One(),
output=synouts.COBA(E=0.), tau=5.,
g_max=g_input['TC'])
self.noise2RTC = synapses.Exponential(self.poisson_RTC, self.RTC, bp.conn.One2One(),
output=synouts.COBA(E=0.), tau=5.,
g_max=g_input['TC'])
self.noise2IN = synapses.Exponential(self.poisson_IN, self.IN, bp.conn.One2One(),
output=synouts.COBA(E=0.), tau=5.,
g_max=g_input['IN'])
self.noise2RE = synapses.Exponential(self.poisson_RE, self.RE, bp.conn.One2One(),
output=synouts.COBA(E=0.), tau=5.,
g_max=g_input['RE'])
# HTC cells were connected with gap junctions
self.gj_HTC = synapses.GapJunction(self.HTC, self.HTC,
bp.conn.ProbDist(dist=2., prob=0.3, ),
comp_method='sparse',
g_max=1e-2)
# HTC provides feedforward excitation to INs
self.HTC2IN_ampa = synapses.AMPA(self.HTC, self.IN, bp.conn.FixedProb(0.3),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
alpha=0.94,
beta=0.18,
g_max=6e-3)
self.HTC2IN_nmda = synapses.AMPA(self.HTC, self.IN, bp.conn.FixedProb(0.3),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=MgBlock(),
alpha=1.,
beta=0.0067,
g_max=3e-3)
# INs delivered feedforward inhibition to RTC cells
self.IN2RTC = synapses.GABAa(self.IN, self.RTC, bp.conn.FixedProb(0.3),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=synouts.COBA(E=-80),
alpha=10.5,
beta=0.166,
g_max=3e-3)
# 20% RTC cells electrically connected with HTC cells
self.gj_RTC2HTC = synapses.GapJunction(self.RTC, self.HTC,
bp.conn.ProbDist(dist=2., prob=0.3, pre_ratio=0.2),
comp_method='sparse',
g_max=1 / 300)
# Both HTC and RTC cells sent glutamatergic synapses to RE neurons, while
# receiving GABAergic feedback inhibition from the RE population
self.HTC2RE_ampa = synapses.AMPA(self.HTC, self.RE, bp.conn.FixedProb(0.2),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
alpha=0.94,
beta=0.18,
g_max=4e-3)
self.RTC2RE_ampa = synapses.AMPA(self.RTC, self.RE, bp.conn.FixedProb(0.2),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
alpha=0.94,
beta=0.18,
g_max=4e-3)
self.HTC2RE_nmda = synapses.AMPA(self.HTC, self.RE, bp.conn.FixedProb(0.2),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=MgBlock(),
alpha=1.,
beta=0.0067,
g_max=2e-3)
self.RTC2RE_nmda = synapses.AMPA(self.RTC, self.RE, bp.conn.FixedProb(0.2),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=MgBlock(),
alpha=1.,
beta=0.0067,
g_max=2e-3)
self.RE2HTC = synapses.GABAa(self.RE, self.HTC, bp.conn.FixedProb(0.2),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=synouts.COBA(E=-80),
alpha=10.5,
beta=0.166,
g_max=3e-3)
self.RE2RTC = synapses.GABAa(self.RE, self.RTC, bp.conn.FixedProb(0.2),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=synouts.COBA(E=-80),
alpha=10.5,
beta=0.166,
g_max=3e-3)
# RE neurons were connected with both gap junctions and GABAergic synapses
self.gj_RE = synapses.GapJunction(self.RE, self.RE,
bp.conn.ProbDist(dist=2., prob=0.3, pre_ratio=0.2),
comp_method='sparse',
g_max=1 / 300)
self.RE2RE = synapses.GABAa(self.RE, self.RE, bp.conn.FixedProb(0.2),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=synouts.COBA(E=-70),
alpha=10.5, beta=0.166,
g_max=1e-3)
# 10% RE neurons project GABAergic synapses to local interneurons
# probability (0.05) was used for the RE->IN synapses according to experimental data
self.RE2IN = synapses.GABAa(self.RE, self.IN, bp.conn.FixedProb(0.05, pre_ratio=0.1),
delay_step=int(2 / bm.get_dt()),
stp=synplast.STD(tau=700, U=0.07),
output=synouts.COBA(E=-80),
alpha=10.5, beta=0.166,
g_max=1e-3, )
Simulation
[8]:
states = {
'delta': dict(g_input={'IN': 1e-4, 'RE': 1e-4, 'TC': 1e-4},
g_KL={'TC': 0.035, 'RE': 0.03, 'IN': 0.01}),
'spindle': dict(g_input={'IN': 3e-4, 'RE': 3e-4, 'TC': 3e-4},
g_KL={'TC': 0.01, 'RE': 0.02, 'IN': 0.015}),
'alpha': dict(g_input={'IN': 1.5e-3, 'RE': 1.5e-3, 'TC': 1.5e-3},
g_KL={'TC': 0., 'RE': 0.01, 'IN': 0.02}),
'gamma': dict(g_input={'IN': 1.5e-3, 'RE': 1.5e-3, 'TC': 1.7e-2},
g_KL={'TC': 0., 'RE': 0.01, 'IN': 0.02}),
}
[9]:
def rhythm_const_input(amp, freq, length, duration, t_start=0., t_end=None, dt=None):
if t_end is None: t_end = duration
if length > duration:
raise ValueError(f'Expected length <= duration, while we got {length} > {duration}')
sec_length = 1e3 / freq
values, durations = [0.], [t_start]
for t in np.arange(t_start, t_end, sec_length):
values.append(amp)
if t + length <= t_end:
durations.append(length)
values.append(0.)
if t + sec_length <= t_end:
durations.append(sec_length - length)
else:
durations.append(t_end - t - length)
else:
durations.append(t_end - t)
values.append(0.)
durations.append(duration - t_end)
return bp.inputs.section_input(values=values, durations=durations, dt=dt, )
[10]:
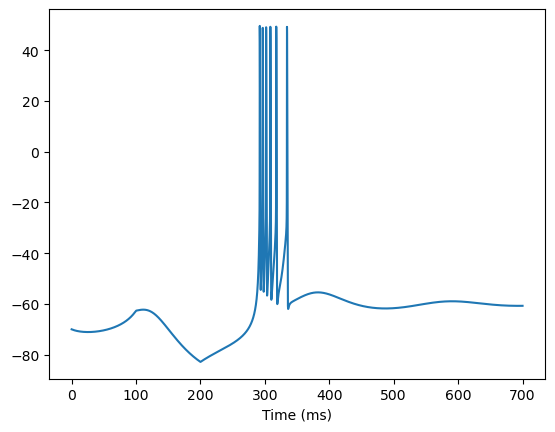
def try_trn_neuron():
trn = TRN(1)
I, length = bp.inputs.section_input(values=[0, -0.05, 0],
durations=[100, 100, 500],
return_length=True,
dt=0.01)
runner = bp.DSRunner(trn,
monitors=['V'],
inputs=['input', I, 'iter'],
dt=0.01)
runner.run(length)
bp.visualize.line_plot(runner.mon.ts, runner.mon.V, show=True)
[11]:
try_trn_neuron()
WARNING:jax._src.lib.xla_bridge:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
[12]:
def try_network(state='delta'):
duration = 3e3
net = Thalamus(
IN_V_init=bp.init.OneInit(-70.),
RE_V_init=bp.init.OneInit(-70.),
HTC_V_init=bp.init.OneInit(-80.),
RTC_V_init=bp.init.OneInit(-80.),
**states[state],
)
net.reset()
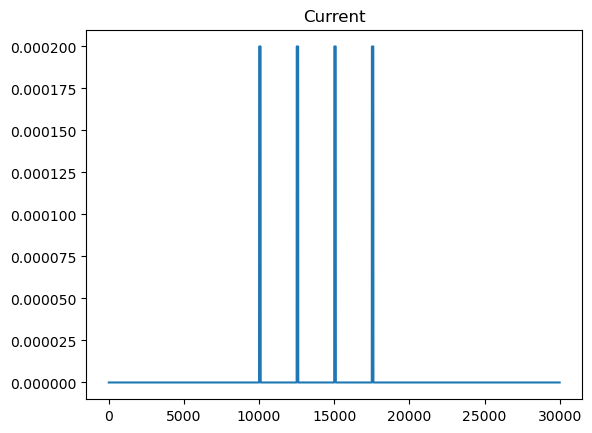
currents = rhythm_const_input(2e-4, freq=4., length=10.,
duration=duration,
t_end=2e3, t_start=1e3)
plt.plot(currents)
plt.title('Current')
plt.show()
runner = bp.DSRunner(
net,
monitors=['HTC.spike', 'RTC.spike', 'RE.spike', 'IN.spike',
'HTC.V', 'RTC.V', 'RE.V', 'IN.V', ],
inputs=[('HTC.input', currents, 'iter'),
('RTC.input', currents, 'iter'),
('IN.input', currents, 'iter')],
)
runner.run(duration)
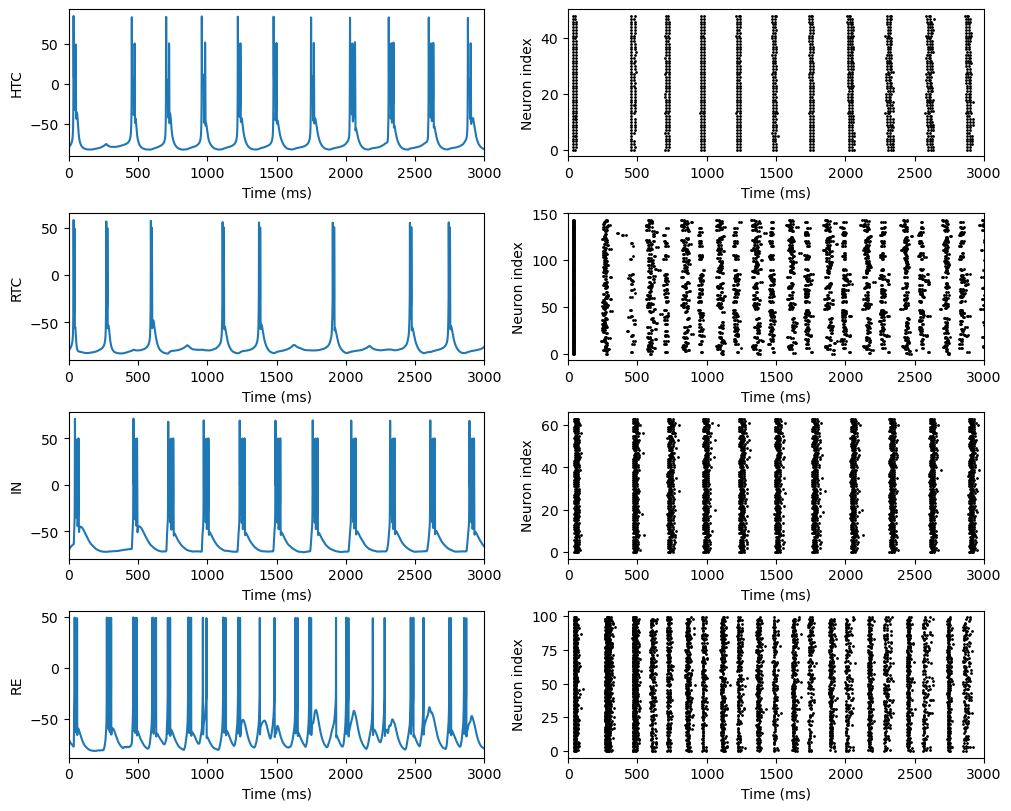
fig, gs = bp.visualize.get_figure(4, 2, 2, 5)
fig.add_subplot(gs[0, 0])
bp.visualize.line_plot(runner.mon.ts, runner.mon.get('HTC.V'), ylabel='HTC', xlim=(0, duration))
fig.add_subplot(gs[1, 0])
bp.visualize.line_plot(runner.mon.ts, runner.mon.get('RTC.V'), ylabel='RTC', xlim=(0, duration))
fig.add_subplot(gs[2, 0])
bp.visualize.line_plot(runner.mon.ts, runner.mon.get('IN.V'), ylabel='IN', xlim=(0, duration))
fig.add_subplot(gs[3, 0])
bp.visualize.line_plot(runner.mon.ts, runner.mon.get('RE.V'), ylabel='RE', xlim=(0, duration))
fig.add_subplot(gs[0, 1])
bp.visualize.raster_plot(runner.mon.ts, runner.mon.get('HTC.spike'), xlim=(0, duration))
fig.add_subplot(gs[1, 1])
bp.visualize.raster_plot(runner.mon.ts, runner.mon.get('RTC.spike'), xlim=(0, duration))
fig.add_subplot(gs[2, 1])
bp.visualize.raster_plot(runner.mon.ts, runner.mon.get('IN.spike'), xlim=(0, duration))
fig.add_subplot(gs[3, 1])
bp.visualize.raster_plot(runner.mon.ts, runner.mon.get('RE.spike'), xlim=(0, duration))
plt.show()
[13]:
try_network()