(Yang, 2020): Dynamical system analysis for RNN
Implementation of the paper:
Yang G R, Wang X J. Artificial neural networks for neuroscientists: A primer[J]. Neuron, 2020, 107(6): 1048-1070.
The original implementation is based on PyTorch: https://github.com/gyyang/nn-brain/blob/master/RNN%2BDynamicalSystemAnalysis.ipynb
[1]:
import brainpy as bp
import brainpy.math as bm
bp.math.set_platform('cpu')
[2]:
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
In this tutorial, we will use supervised learning to train a recurrent neural network on a simple perceptual decision making task, and analyze the trained network using dynamical system analysis.
Defining a cognitive task
[3]:
# We will import the task from the neurogym library.
# Please install neurogym:
#
# https://github.com/neurogym/neurogym
import neurogym as ngym
[4]:
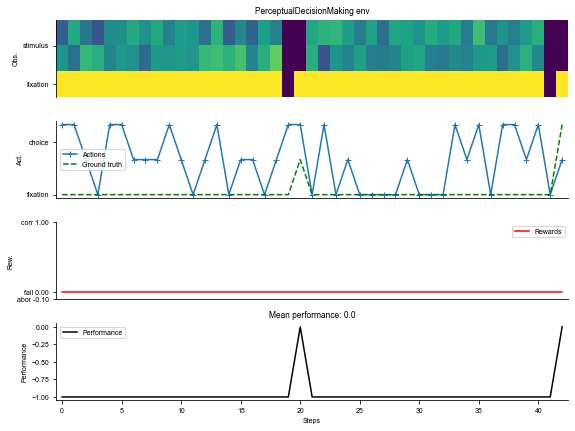
# Environment
task = 'PerceptualDecisionMaking-v0'
kwargs = {'dt': 100}
seq_len = 100
# Make supervised dataset
dataset = ngym.Dataset(task, env_kwargs=kwargs, batch_size=16,
seq_len=seq_len)
# A sample environment from dataset
env = dataset.env
# Visualize the environment with 2 sample trials
_ = ngym.utils.plot_env(env, num_trials=2, fig_kwargs={'figsize': (8, 6)})
[5]:
input_size = env.observation_space.shape[0]
output_size = env.action_space.n
batch_size = dataset.batch_size
Define a vanilla continuous-time recurrent network
Here we will define a continuous-time neural network but discretize it in time using the Euler method.
This continuous-time system can then be discretized using the Euler method with a time step of \(\Delta t\),
[6]:
class RNN(bp.DynamicalSystem):
def __init__(self,
num_input,
num_hidden,
num_output,
num_batch,
dt=None, seed=None,
w_ir=bp.init.KaimingNormal(scale=1.),
w_rr=bp.init.KaimingNormal(scale=1.),
w_ro=bp.init.KaimingNormal(scale=1.)):
super(RNN, self).__init__()
# parameters
self.tau = 100
self.num_batch = num_batch
self.num_input = num_input
self.num_hidden = num_hidden
self.num_output = num_output
if dt is None:
self.alpha = 1
else:
self.alpha = dt / self.tau
self.rng = bm.random.RandomState(seed=seed)
# input weight
self.w_ir = bm.TrainVar(bp.init.parameter(w_ir, (num_input, num_hidden)))
# recurrent weight
bound = 1 / num_hidden ** 0.5
self.w_rr = bm.TrainVar(bp.init.parameter(w_rr, (num_hidden, num_hidden)))
self.b_rr = bm.TrainVar(self.rng.uniform(-bound, bound, num_hidden))
# readout weight
self.w_ro = bm.TrainVar(bp.init.parameter(w_ro, (num_hidden, num_output)))
self.b_ro = bm.TrainVar(self.rng.uniform(-bound, bound, num_output))
# variables
self.h = bm.Variable(bm.zeros((num_batch, num_hidden)))
self.o = bm.Variable(bm.zeros((num_batch, num_output)))
def cell(self, x, h):
ins = x @ self.w_ir + h @ self.w_rr + self.b_rr
state = h * (1 - self.alpha) + ins * self.alpha
return bm.relu(state)
def readout(self, h):
return h @ self.w_ro + self.b_ro
def update(self, x):
self.h.value = self.cell(x, self.h)
self.o.value = self.readout(self.h)
return self.h.value, self.o.value
def predict(self, xs):
self.h[:] = 0.
return bm.for_loop(self.update, xs, dyn_vars=[self.h, self.o])
def loss(self, xs, ys):
hs, os = self.predict(xs)
os = os.reshape((-1, os.shape[-1]))
loss = bp.losses.cross_entropy_loss(os, ys.flatten())
return loss, os
Train the recurrent network on the decision-making task
[7]:
# Instantiate the network and print information
hidden_size = 64
with bm.training_environment():
net = RNN(num_input=input_size,
num_hidden=hidden_size,
num_output=output_size,
num_batch=batch_size,
dt=env.dt)
[8]:
# Adam optimizer
opt = bp.optim.Adam(lr=0.001, train_vars=net.train_vars().unique())
# gradient function
grad_f = bm.grad(net.loss,
child_objs=net,
grad_vars=net.train_vars().unique(),
return_value=True,
has_aux=True)
# training function
@bm.jit
@bm.to_object(child_objs=(grad_f, opt))
def train(xs, ys):
grads, loss, os = grad_f(xs, ys)
opt.update(grads)
return loss, os
[9]:
running_acc = 0
running_loss = 0
for i in range(1500):
inputs, labels_np = dataset()
inputs = bm.asarray(inputs)
labels = bm.asarray(labels_np)
loss, outputs = train(inputs, labels)
running_loss += loss
# Compute performance
output_np = np.argmax(bm.as_numpy(outputs), axis=-1).flatten()
labels_np = labels_np.flatten()
ind = labels_np > 0 # Only analyze time points when target is not fixation
running_acc += np.mean(labels_np[ind] == output_np[ind])
if i % 100 == 99:
running_loss /= 100
running_acc /= 100
print('Step {}, Loss {:0.4f}, Acc {:0.3f}'.format(i + 1, running_loss, running_acc))
running_loss = 0
running_acc = 0
Step 100, Loss 0.0994, Acc 0.458
Step 200, Loss 0.0201, Acc 0.868
Step 300, Loss 0.0155, Acc 0.876
Step 400, Loss 0.0136, Acc 0.883
Step 500, Loss 0.0129, Acc 0.878
Step 600, Loss 0.0127, Acc 0.881
Step 700, Loss 0.0122, Acc 0.884
Step 800, Loss 0.0123, Acc 0.884
Step 900, Loss 0.0116, Acc 0.885
Step 1000, Loss 0.0116, Acc 0.885
Step 1100, Loss 0.0111, Acc 0.888
Step 1200, Loss 0.0111, Acc 0.885
Step 1300, Loss 0.0108, Acc 0.891
Step 1400, Loss 0.0111, Acc 0.885
Step 1500, Loss 0.0106, Acc 0.891
Visualize neural activity for in sample trials
We will run the network for 100 sample trials, then visual the neural activity trajectories in a PCA space.
[10]:
env.reset(no_step=True)
perf = 0
num_trial = 100
activity_dict = {}
trial_infos = {}
for i in range(num_trial):
env.new_trial()
ob, gt = env.ob, env.gt
inputs = bm.asarray(ob[:, np.newaxis, :])
rnn_activity, action_pred = net.predict(inputs)
rnn_activity = bm.as_numpy(rnn_activity)[:, 0, :]
activity_dict[i] = rnn_activity
trial_infos[i] = env.trial
# Concatenate activity for PCA
activity = np.concatenate(list(activity_dict[i] for i in range(num_trial)), axis=0)
print('Shape of the neural activity: (Time points, Neurons): ', activity.shape)
# Print trial informations
for i in range(5):
print('Trial ', i, trial_infos[i])
Shape of the neural activity: (Time points, Neurons): (2200, 64)
Trial 0 {'ground_truth': 0, 'coh': 25.6}
Trial 1 {'ground_truth': 1, 'coh': 0.0}
Trial 2 {'ground_truth': 0, 'coh': 6.4}
Trial 3 {'ground_truth': 0, 'coh': 6.4}
Trial 4 {'ground_truth': 1, 'coh': 0.0}
[11]:
pca = PCA(n_components=2)
pca.fit(activity)
[11]:
PCA(n_components=2)
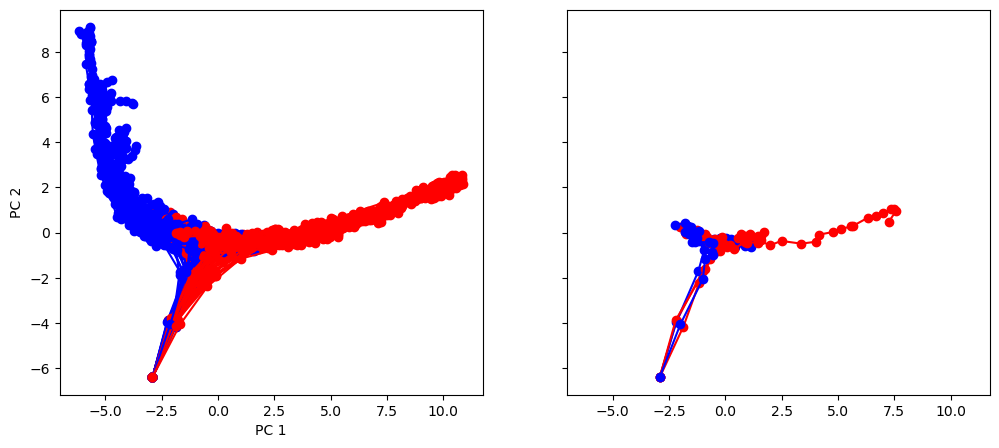
Transform individual trials and Visualize in PC space based on ground-truth color. We see that the neural activity is organized by stimulus ground-truth in PC1
[12]:
plt.rcdefaults()
fig, (ax1, ax2) = plt.subplots(1, 2, sharey=True, sharex=True, figsize=(12, 5))
for i in range(num_trial):
activity_pc = pca.transform(activity_dict[i])
trial = trial_infos[i]
color = 'red' if trial['ground_truth'] == 0 else 'blue'
_ = ax1.plot(activity_pc[:, 0], activity_pc[:, 1], 'o-', color=color)
if i < 5:
_ = ax2.plot(activity_pc[:, 0], activity_pc[:, 1], 'o-', color=color)
ax1.set_xlabel('PC 1')
ax1.set_ylabel('PC 2')
plt.show()
Search for approximate fixed points
Here we search for approximate fixed points and visualize them in the same PC space. In a generic dynamical system,
We can search for fixed points by doing the optimization
[13]:
f_cell = lambda h: net.cell(bm.asarray([1, 0.5, 0.5]), h)
[14]:
fp_candidates = bm.vstack([activity_dict[i] for i in range(num_trial)])
fp_candidates.shape
[14]:
(2200, 64)
[15]:
finder = bp.analysis.SlowPointFinder(f_cell=f_cell, f_type='discrete')
finder.find_fps_with_gd_method(
candidates=fp_candidates,
tolerance=1e-5, num_batch=200,
optimizer=bp.optim.Adam(lr=bp.optim.ExponentialDecay(0.01, 1, 0.9999)),
)
finder.filter_loss(tolerance=1e-5)
finder.keep_unique(tolerance=0.03)
finder.exclude_outliers(0.1)
fixed_points = finder.fixed_points
Optimizing with Adam(lr=ExponentialDecay(0.01, decay_steps=1, decay_rate=0.9999), last_call=-1), beta1=0.9, beta2=0.999, eps=1e-08) to find fixed points:
Batches 1-200 in 0.74 sec, Training loss 0.0012789472
Batches 201-400 in 0.69 sec, Training loss 0.0005247793
Batches 401-600 in 0.68 sec, Training loss 0.0003411663
Batches 601-800 in 0.69 sec, Training loss 0.0002612533
Batches 801-1000 in 0.69 sec, Training loss 0.0002112046
Batches 1001-1200 in 0.73 sec, Training loss 0.0001740992
Batches 1201-1400 in 0.70 sec, Training loss 0.0001455246
Batches 1401-1600 in 0.74 sec, Training loss 0.0001233126
Batches 1601-1800 in 0.70 sec, Training loss 0.0001058684
Batches 1801-2000 in 0.77 sec, Training loss 0.0000921902
Batches 2001-2200 in 0.72 sec, Training loss 0.0000814899
Batches 2201-2400 in 0.57 sec, Training loss 0.0000731918
Batches 2401-2600 in 0.75 sec, Training loss 0.0000667152
Batches 2601-2800 in 0.74 sec, Training loss 0.0000616670
Batches 2801-3000 in 0.70 sec, Training loss 0.0000576638
Batches 3001-3200 in 0.56 sec, Training loss 0.0000543614
Batches 3201-3400 in 0.60 sec, Training loss 0.0000515535
Batches 3401-3600 in 0.61 sec, Training loss 0.0000491180
Batches 3601-3800 in 0.71 sec, Training loss 0.0000469084
Batches 3801-4000 in 0.75 sec, Training loss 0.0000448209
Batches 4001-4200 in 0.67 sec, Training loss 0.0000428211
Batches 4201-4400 in 0.73 sec, Training loss 0.0000408868
Batches 4401-4600 in 0.71 sec, Training loss 0.0000390110
Batches 4601-4800 in 0.77 sec, Training loss 0.0000371877
Batches 4801-5000 in 0.58 sec, Training loss 0.0000354225
Batches 5001-5200 in 0.58 sec, Training loss 0.0000337130
Batches 5201-5400 in 0.69 sec, Training loss 0.0000320665
Batches 5401-5600 in 0.65 sec, Training loss 0.0000304907
Batches 5601-5800 in 0.76 sec, Training loss 0.0000289806
Batches 5801-6000 in 0.64 sec, Training loss 0.0000275472
Batches 6001-6200 in 0.55 sec, Training loss 0.0000261840
Batches 6201-6400 in 0.60 sec, Training loss 0.0000248957
Batches 6401-6600 in 0.68 sec, Training loss 0.0000236831
Batches 6601-6800 in 0.67 sec, Training loss 0.0000225569
Batches 6801-7000 in 0.71 sec, Training loss 0.0000215175
Batches 7001-7200 in 0.66 sec, Training loss 0.0000205547
Batches 7201-7400 in 0.70 sec, Training loss 0.0000196768
Batches 7401-7600 in 0.68 sec, Training loss 0.0000188892
Batches 7601-7800 in 0.71 sec, Training loss 0.0000181973
Batches 7801-8000 in 0.67 sec, Training loss 0.0000175776
Batches 8001-8200 in 0.72 sec, Training loss 0.0000170112
Batches 8201-8400 in 0.75 sec, Training loss 0.0000165215
Batches 8401-8600 in 0.54 sec, Training loss 0.0000160938
Batches 8601-8800 in 0.60 sec, Training loss 0.0000157249
Batches 8801-9000 in 0.52 sec, Training loss 0.0000154070
Batches 9001-9200 in 0.68 sec, Training loss 0.0000151405
Batches 9201-9400 in 0.54 sec, Training loss 0.0000149151
Batches 9401-9600 in 0.53 sec, Training loss 0.0000147287
Batches 9601-9800 in 0.53 sec, Training loss 0.0000145786
Batches 9801-10000 in 0.53 sec, Training loss 0.0000144657
Excluding fixed points with squared speed above tolerance 1e-05:
Kept 1157/2200 fixed points with tolerance under 1e-05.
Excluding non-unique fixed points:
Kept 10/1157 unique fixed points with uniqueness tolerance 0.03.
Excluding outliers:
Kept 8/10 fixed points with within outlier tolerance 0.1.
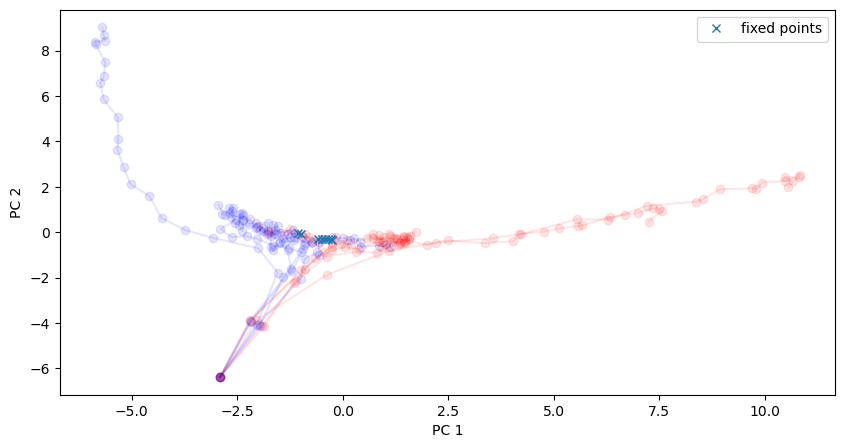
Visualize the found approximate fixed points.
We see that they found an approximate line attrator, corresponding to our PC1, along which evidence is integrated during the stimulus period.
[16]:
# Plot in the same space as activity
plt.figure(figsize=(10, 5))
for i in range(10):
activity_pc = pca.transform(activity_dict[i])
trial = trial_infos[i]
color = 'red' if trial['ground_truth'] == 0 else 'blue'
plt.plot(activity_pc[:, 0], activity_pc[:, 1], 'o-', color=color, alpha=0.1)
# Fixed points are shown in cross
fixedpoints_pc = pca.transform(fixed_points)
plt.plot(fixedpoints_pc[:, 0], fixedpoints_pc[:, 1], 'x', label='fixed points')
plt.xlabel('PC 1')
plt.ylabel('PC 2')
plt.legend()
plt.show()
Computing the Jacobian and finding the line attractor
[17]:
from jax import jacobian
[19]:
dFdh = jacobian(f_cell)(fixed_points[2])
eigval, eigvec = np.linalg.eig(bm.as_numpy(dFdh))
[20]:
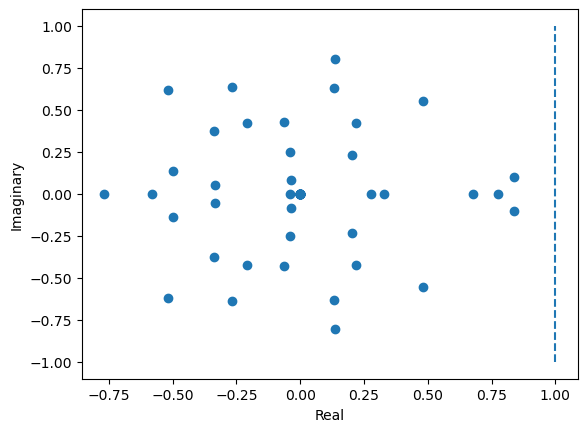
# Plot distribution of eigenvalues in a 2-d real-imaginary plot
plt.figure()
plt.scatter(np.real(eigval), np.imag(eigval))
plt.plot([1, 1], [-1, 1], '--')
plt.xlabel('Real')
plt.ylabel('Imaginary')
plt.show()