(Brette, et, al., 2007) COBA
Implementation of the paper:
Brette, R., Rudolph, M., Carnevale, T., Hines, M., Beeman, D., Bower, J. M., et al. (2007), Simulation of networks of spiking neurons: a review of tools and strategies., J. Comput. Neurosci., 23, 3, 349–98
which is based on the balanced network proposed by:
Vogels, T. P. and Abbott, L. F. (2005), Signal propagation and logic gating in networks of integrate-and-fire neurons., J. Neurosci., 25, 46, 10786–95
Authors:
[1]:
import brainpy as bp
import brainpy.math as bm
bp.math.set_platform('cpu')
[2]:
bp.__version__
[2]:
'2.4.4.post1'
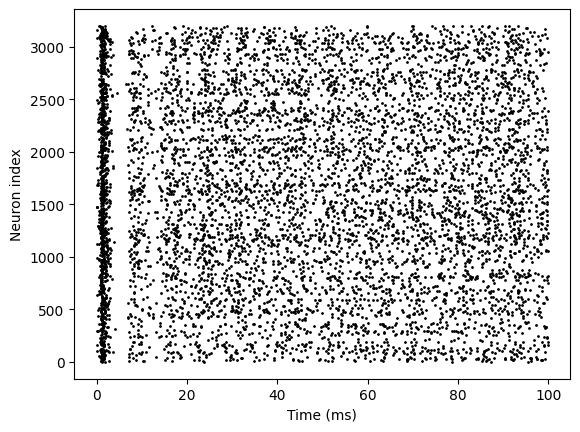
Version 1
[3]:
class EINet(bp.Network):
def __init__(self, scale=1.0, method='exp_auto'):
# network size
num_exc = int(3200 * scale)
num_inh = int(800 * scale)
# neurons
pars = dict(V_rest=-60., V_th=-50., V_reset=-60., tau=20., tau_ref=5.,
V_initializer=bp.init.Normal(-55., 2.))
E = bp.neurons.LIF(num_exc, **pars, method=method)
I = bp.neurons.LIF(num_inh, **pars, method=method)
# synapses
we = 0.6 / scale # excitatory synaptic weight (voltage)
wi = 6.7 / scale # inhibitory synaptic weight
E2E = bp.synapses.Exponential(E, E, bp.conn.FixedProb(prob=0.02),
g_max=we, tau=5., method=method,
output=bp.synouts.COBA(E=0.))
E2I = bp.synapses.Exponential(E, I, bp.conn.FixedProb(prob=0.02),
g_max=we, tau=5., method=method,
output=bp.synouts.COBA(E=0.))
I2E = bp.synapses.Exponential(I, E, bp.conn.FixedProb(prob=0.02),
g_max=wi, tau=10., method=method,
output=bp.synouts.COBA(E=-80.))
I2I = bp.synapses.Exponential(I, I, bp.conn.FixedProb(prob=0.02),
g_max=wi, tau=10., method=method,
output=bp.synouts.COBA(E=-80.))
super(EINet, self).__init__(E2E, E2I, I2E, I2I, E=E, I=I)
[4]:
# network
net = EINet()
[5]:
# simulation
runner = bp.DSRunner(
net,
monitors=['E.spike'],
inputs=[('E.input', 20.), ('I.input', 20.)]
)
runner.run(100.)
[6]:
bp.visualize.raster_plot(runner.mon.ts, runner.mon['E.spike'], show=True)
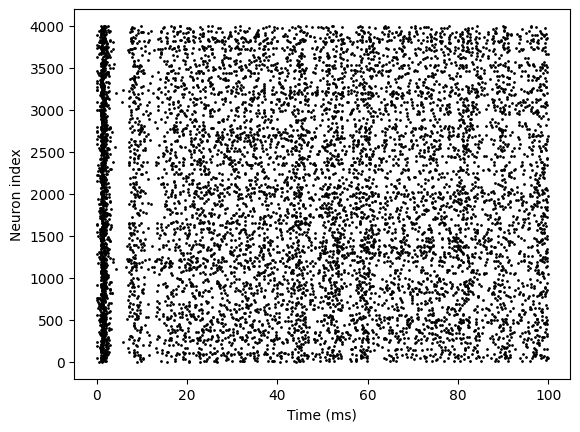
Version 2
[7]:
class EINet_V2(bp.Network):
def __init__(self, scale=1.0, method='exp_auto'):
super(EINet_V2, self).__init__()
# network size
num_exc = int(3200 * scale)
num_inh = int(800 * scale)
# neurons
self.N = bp.neurons.LIF(num_exc + num_inh,
V_rest=-60., V_th=-50., V_reset=-60., tau=20., tau_ref=5.,
method=method, V_initializer=bp.initialize.Normal(-55., 2.))
# synapses
we = 0.6 / scale # excitatory synaptic weight (voltage)
wi = 6.7 / scale # inhibitory synaptic weight
self.Esyn = bp.synapses.Exponential(pre=self.N[:num_exc],
post=self.N,
conn=bp.connect.FixedProb(0.02),
g_max=we, tau=5.,
output=bp.synouts.COBA(E=0.),
method=method)
self.Isyn = bp.synapses.Exponential(pre=self.N[num_exc:],
post=self.N,
conn=bp.connect.FixedProb(0.02),
g_max=wi, tau=10.,
output=bp.synouts.COBA(E=-80.),
method=method)
[8]:
net = EINet_V2(scale=1., method='exp_auto')
# simulation
runner = bp.DSRunner(
net,
monitors={'spikes': net.N.spike},
inputs=[(net.N.input, 20.)]
)
runner.run(100.)
# visualization
bp.visualize.raster_plot(runner.mon.ts, runner.mon['spikes'], show=True)
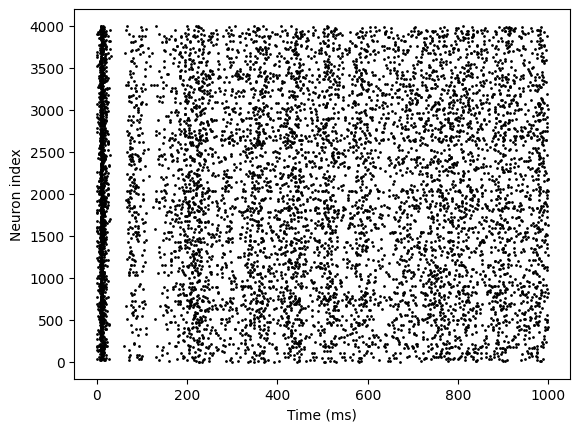
New Version
[9]:
class EINet3(bp.DynSysGroup):
def __init__(self):
super().__init__()
self.N = bp.dyn.LifRef(4000, V_rest=-60., V_th=-50., V_reset=-60., tau=20., tau_ref=5.,
V_initializer=bp.init.Normal(-55., 2.))
self.delay = bp.VarDelay(self.N.spike, entries={'I': None})
self.E = bp.dyn.ProjAlignPostMg1(comm=bp.dnn.EventJitFPHomoLinear(3200, 4000, prob=0.02, weight=0.6),
syn=bp.dyn.Expon.desc(size=4000, tau=5.),
out=bp.dyn.COBA.desc(E=0.),
post=self.N)
self.I = bp.dyn.ProjAlignPostMg1(comm=bp.dnn.EventJitFPHomoLinear(800, 4000, prob=0.02, weight=6.7),
syn=bp.dyn.Expon.desc(size=4000, tau=10.),
out=bp.dyn.COBA.desc(E=-80.),
post=self.N)
def update(self, input):
spk = self.delay.at('I')
self.E(spk[:3200])
self.I(spk[3200:])
self.delay(self.N(input))
return self.N.spike.value
[10]:
model = EINet3()
indices = bm.arange(1000)
spks = bm.for_loop(lambda i: model.step_run(i, 20.), indices)
bp.visualize.raster_plot(indices, spks, show=True)