(Vreeswijk & Sompolinsky, 1996) E/I balanced network
Overviews
Van Vreeswijk and Sompolinsky proposed E-I balanced network in 1996 to explain the temporally irregular spiking patterns. They suggested that the temporal variability may originated from the balance between excitatory and inhibitory inputs.
There are \(N_E\) excitatory neurons and \(N_I\) inbibitory neurons.
An important feature of the network is random and sparse connectivity. Connections between neurons \(K\) meets \(1 << K << N_E\).
Implementations
[13]:
import brainpy as bp
import brainpy.math as bm
bm.set_platform('cpu')
[14]:
bp.__version__
[14]:
'2.4.4.post1'
Dynamic of membrane potential is given as:
where \(I_i^{net}(t)\) represents the synaptic current, which describes the sum of excitatory and inhibitory neurons.
where
Parameters: \(J_E = \frac 1 {\sqrt {pN_e}}, J_I = \frac 1 {\sqrt {pN_i}}\)
We can see from the dynamic that network is based on leaky Integrate-and-Fire neurons, and we can just use get_LIF from bpmodels.neurons to get this model.
The function of \(I_i^{net}(t)\) is actually a synase with single exponential decay, we can also get it by using get_exponential.
Synapse
[15]:
class ExponCUBA(bp.Projection):
def __init__(self, pre, post, prob, g_max, tau):
super().__init__()
self.proj = bp.dyn.ProjAlignPostMg2(
pre=pre,
delay=None,
comm=bp.dnn.EventCSRLinear(bp.conn.FixedProb(prob, pre=pre.num, post=post.num), g_max),
syn=bp.dyn.Expon.desc(post.num, tau=tau),
out=bp.dyn.CUBA.desc(),
post=post,
)
Network
Let’s create a neuron group with \(N_E\) excitatory neurons and \(N_I\) inbibitory neurons. Use conn=bp.connect.FixedProb(p) to implement the random and sparse connections.
[16]:
class EINet(bp.DynSysGroup):
def __init__(self, num_exc, num_inh, prob, JE, JI):
# neurons
pars = dict(V_rest=-52., V_th=-50., V_reset=-60., tau=10., tau_ref=0.,
V_initializer=bp.init.Normal(-60., 10.))
E = bp.neurons.LIF(num_exc, **pars)
I = bp.neurons.LIF(num_inh, **pars)
# synapses
E2E = ExponCUBA(E, E, prob, g_max=JE, tau=2.)
E2I = ExponCUBA(E, I, prob, g_max=JE, tau=2.)
I2E = ExponCUBA(I, E, prob, g_max=JI, tau=2.)
I2I = ExponCUBA(I, I, prob, g_max=JI, tau=2.)
super(EINet, self).__init__(E2E, E2I, I2E, I2I, E=E, I=I)
[17]:
num_exc = 500
num_inh = 500
prob = 0.1
Ib = 3.
JE = 1 / bp.math.sqrt(prob * num_exc)
JI = -1 / bp.math.sqrt(prob * num_inh)
[18]:
net = EINet(num_exc, num_inh, prob=prob, JE=JE, JI=JI)
runner = bp.DSRunner(net,
monitors=['E.spike'],
inputs=[('E.input', Ib), ('I.input', Ib)])
t = runner.run(1000.)
Visualization
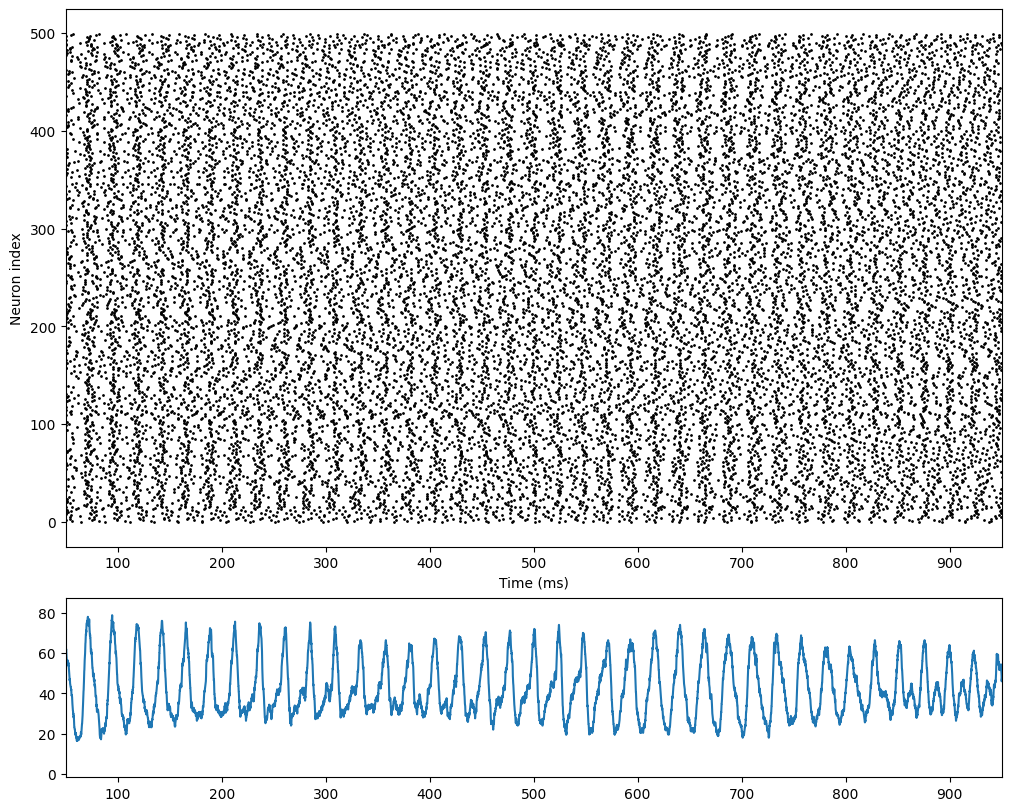
[19]:
import matplotlib.pyplot as plt
fig, gs = bp.visualize.get_figure(4, 1, 2, 10)
fig.add_subplot(gs[:3, 0])
bp.visualize.raster_plot(runner.mon.ts, runner.mon['E.spike'], xlim=(50, 950))
fig.add_subplot(gs[3, 0])
rates = bp.measure.firing_rate(runner.mon['E.spike'], 5.)
plt.plot(runner.mon.ts, rates)
plt.xlim(50, 950)
plt.show()
Reference
[1] Van Vreeswijk, Carl, and Haim Sompolinsky. “Chaos in neuronal networks with balanced excitatory and inhibitory activity.” Science 274.5293 (1996): 1724-1726.
