(Joglekar, et. al, 2018): Inter-areal Balanced Amplification taichi customized operators
Using taichi customized operators to implement the model in (Joglekar, et. al, 2018).:
Joglekar, Madhura R., et al. “Inter-areal balanced amplification enhances signal propagation in a large-scale circuit model of the primate cortex.” Neuron 98.1 (2018): 222-234.
[17]:
import brainpy as bp
import brainpy.math as bm
from brainpy import neurons
import jax
import jax.numpy as jnp
import matplotlib.pyplot as plt
import numpy as np
from scipy.io import loadmat
from typing import Tuple, Optional
from functools import partial
[18]:
bp.__version__
[18]:
'2.5.0'
You should import taichi before customize your operators
[19]:
import taichi as ti
And there are some random number generators which were defined in BrainPy, We need to use these in taichi kernels
[20]:
from brainpy.math.tifunc import (lfsr88_key, lfsr88_random_integers)
You can select the target platform you want to run your customized operators
[21]:
# bm.set_platform('cpu')
bm.set_platform('gpu')
Define the kernel with taichi.kernel decorater
For customized your operators, you can find the tutorial at CPU and GPU Operator Customization with Taichi
Below is defining a Taichi kernel for processing multiple areas with event-driven updates.
This kernel receive spikes from the excitory neurons of one region and calculate the output currents to the neurons of all regions.
[22]:
@ti.kernel
def _multiple_area_event_mv_prob_homo_irregular_cpu_E(
events: ti.types.ndarray(ndim=1), # 1D array of events, where nonzero values indicate a spike.
weight: ti.types.ndarray(ndim=1), # 1D array of weights, one per area, to be added to the output on an event.
clen: ti.types.ndarray(ndim=1), # 1D array containing the upper limit for random integer generation.
seed: ti.types.ndarray(ndim=1), # 1D array of seeds, one per area, for random number generation.
out: ti.types.ndarray(ndim=2) # 2D output array where current updates are made based on events and weights.
):
num_row = out.shape[1] # Number of rows in the output array.
num_col = events.shape[0] # Number of columns (events) to process.
clen0 = clen[0] # Upper limit for random integer generation.
for i_col in range(num_col): # Iterate over each event.
if events[i_col] != 0.: # Check if the current event occurred (nonzero).
for i_area in range(out.shape[0]): # Iterate over each area.
key = lfsr88_key(seed[i_area] + i_col) # Generate an initial key for random numbers.
key, i_row = lfsr88_random_integers(key, 0, clen0 - 1) # Generate the first random row index.
while i_row < num_row: # Ensure the random index is within bounds.
out[i_area, i_row] += weight[i_area] # Update the output array with the weight.
key, inc = lfsr88_random_integers(key, 1, clen0) # Generate the next random increment.
i_row += inc # Update the row index based on the random increment.
For the GPU version, almost as same above kernel. The only difference is that we use ti.ndrange to merge two for loops into one for loop in order to improve the parallelism on GPU.
[23]:
@ti.kernel
def _multiple_area_event_mv_prob_homo_irregular_gpu_E(
events: ti.types.ndarray(ndim=1),
weight: ti.types.ndarray(ndim=1),
clen: ti.types.ndarray(ndim=1),
seed: ti.types.ndarray(ndim=1),
out: ti.types.ndarray(ndim=2)
):
num_row = out.shape[1]
num_col = events.shape[0]
clen0 = clen[0]
area_num = out.shape[0]
# Due to the arch of GPU, use ti.ndrange for parallel processing across events and areas.
for i_col, i_area in ti.ndrange(num_col, area_num):
if events[i_col] != 0.:
key = lfsr88_key(seed[i_area] + i_col)
key, i_row = lfsr88_random_integers(key, 0, clen0 - 1)
while i_row < num_row:
out[i_area, i_row] += weight[i_area]
key, inc = lfsr88_random_integers(key, 1, clen0)
i_row += inc
This kernel receive spikes from the inhibitory neurons of all region and calculate the output currents to the neurons of all regions. (Inhibitory neurons here only affect neurons in their own brain region)
[24]:
@ti.kernel
def _multiple_area_event_mv_prob_homo_irregular_cpu_I(
events: ti.types.ndarray(ndim=2), # 2D array of events, where nonzero values indicate a spike in a region .
weight: ti.types.ndarray(ndim=1), # 1D array of weights, one per area, to be added to the output on an event.
clen: ti.types.ndarray(ndim=1), # 1D array containing the upper limit for random integer generation.
seed: ti.types.ndarray(ndim=1), # 1D array of seeds, one per area, for random number generation.
out: ti.types.ndarray(ndim=2) # 2D output array where current updates are made based on events and weights.
):
num_row = out.shape[1] # Number of rows in the output array.
num_col = events.shape[1] # Number of columns (events) to process.
clen0 = clen[0] # Upper limit for random integer generation.
weight0 = weight[0] # Each area has same weight
for i_col in range(num_col): # Iterate over each event.
for i_area in range(out.shape[0]): # Inhibitory neurons affect only those in their own region
if events[i_area, i_col] != 0.: # Check if the current event occurred (nonzero).
key = lfsr88_key(seed[i_area] + i_col) # Generate an initial key for random numbers.
key, i_row = lfsr88_random_integers(key, 0, clen0 - 1) # Generate the first random row index.
while i_row < num_row: # Ensure the random index is within bounds.
out[i_area, i_row] += weight0 # Update the output array with the weight.
key, inc = lfsr88_random_integers(key, 1, clen0) # Generate the next random increment.
i_row += inc # Update the row index based on the random increment.
For the GPU version, almost as same above kernel. The only difference is that we use ti.ndrange to merge two for loops into one for loop in order to improve the parallelism on GPU.
[25]:
@ti.kernel
def _multiple_area_event_mv_prob_homo_irregular_gpu_I(
events: ti.types.ndarray(ndim=1),
weight: ti.types.ndarray(ndim=1),
clen: ti.types.ndarray(ndim=1),
seed: ti.types.ndarray(ndim=1),
out: ti.types.ndarray(ndim=2)
):
num_row = out.shape[1]
num_col = events.shape[1]
weight0 = weight[0]
clen0 = clen[0]
area_num = out.shape[0]
# Due to the arch of GPU, use ti.ndrange for parallel processing across events and areas.
for i_col, i_area in ti.ndrange(num_col, area_num):
if events[i_area, i_col] != 0.:
key = lfsr88_key(seed[i_area] + i_col)
key, i_row = lfsr88_random_integers(key, 0, clen0 - 1)
while i_row < num_row:
out[i_area, i_row] += weight0
key, inc = lfsr88_random_integers(key, 1, clen0)
i_row += inc
Register the kernel to BrainPy by bm.XLACustomOp
[26]:
_multiple_area_event_mv_prob_homo_irregular_p_E = bm.XLACustomOp(cpu_kernel=_multiple_area_event_mv_prob_homo_irregular_cpu_E,
gpu_kernel=_multiple_area_event_mv_prob_homo_irregular_gpu_E)
_multiple_area_event_mv_prob_homo_irregular_p_I = bm.XLACustomOp(cpu_kernel=_multiple_area_event_mv_prob_homo_irregular_cpu_I,
gpu_kernel=_multiple_area_event_mv_prob_homo_irregular_gpu_I)
Callable apis for further encapsulation
This API receive spikes from the excitory neurons of one region and calculate the output currents to the neurons of all regions.
[27]:
def multiple_area_customized_op_E(
events: jax.Array, # Input events, expected to be a 1D array.
weight: float, # Uniform weight for all updates.
seed: list[int], # List of seeds for random number generation of each region
conn_prob: float, # Connection probability between events and areas.
*,
shape: Tuple[int, int], # Shape of the output matrix.
area_num: int, # Number of areas (rows in the output matrix).
):
# Convert inputs to JAX arrays for compatibility.
events = bm.as_jax(events)
if isinstance(weight, float): weight = bm.as_jax(weight)
weight = jnp.atleast_1d(bm.as_jax(weight))
# Calculate the connection length based on connection probability.
conn_len = jnp.ceil(1 / conn_prob) * 2 - 1
conn_len = jnp.asarray(jnp.atleast_1d(conn_len), dtype=jnp.int32)
# Define the shape of the output matrix and verify shape compatibility.
out_shape = (area_num, shape[1])
if events.shape[0] != shape[0]:
raise ValueError(f'Shape mismatch, vec {events.shape} @ mat {shape}.')
shape = shape[::-1]
# Call the primitive operation
return _multiple_area_event_mv_prob_homo_irregular_p_E(events,
weight,
conn_len,
seed,
outs=[jax.ShapeDtypeStruct(shape=out_shape, dtype=weight.dtype)],
shape=shape,
area_num=area_num)[0]
This API receive spikes from the inhibitory neurons of all region and calculate the output currents to the neurons of all regions. (Inhibitory neurons here only affect neurons in their own brain region)
[28]:
def multiple_area_customized_op_I(
events: jax.Array, # Input events, expected to be a 2D array.
weight: float, # Uniform weight for all updates.
seed: list[int], # List of seeds for random number generation of each region
conn_prob: float, # Connection probability.
*,
shape: Tuple[int, int], # Shape of the output matrix.
area_num: int, # Number of areas.
):
# Similar preparation and validation steps as in multiple_area_customized_op_E.
events = bm.as_jax(events)
if isinstance(weight, float): weight = bm.as_jax(weight)
weight = jnp.atleast_1d(bm.as_jax(weight))
conn_len = jnp.ceil(1 / conn_prob) * 2 - 1
conn_len = jnp.asarray(jnp.atleast_1d(conn_len), dtype=jnp.int32)
# Adjusts shape and calls the primitive operation with validated inputs.
out_shape = (area_num, shape[1])
if events.shape[1] != shape[0]:
raise ValueError(f'Shape mismatch, vec {events.shape} @ mat {shape}.')
shape = shape[::-1]
# Call the primitive operation
return _multiple_area_event_mv_prob_homo_irregular_p_I(events,
weight,
conn_len,
seed,
outs=[jax.ShapeDtypeStruct(shape=out_shape, dtype=weight.dtype)],
shape=shape,
area_num=area_num)[0]
Define Multiple Area Network
[29]:
class MultiAreaNet(bp.Network):
def __init__(
self, hier, conn, delay_mat, muIE=0.0475, muEE=.0375, wII=.075,
wEE=.01, wIE=.075, wEI=.0375, extE=15.4, extI=14.0, alpha=4., seed=None,
):
super(MultiAreaNet, self).__init__()
# data
self.hier = hier
self.conn = conn
self.delay_mat = delay_mat
# parameters
self.muIE = muIE
self.muEE = muEE
self.wII = wII
self.wEE = wEE
self.wIE = wIE
self.wEI = wEI
self.extE = extE
self.extI = extI
self.alpha = alpha
num_area = hier.size
self.num_area = num_area
# neuron models
self.E = neurons.LIF((num_area, 1600),
V_th=-50., V_reset=-60.,
V_rest=-70., tau=20., tau_ref=2.,
noise=3. / bm.sqrt(20.),
V_initializer=bp.init.Uniform(-70., -50.),
method='exp_auto',
keep_size=True,
ref_var=True)
self.I = neurons.LIF((num_area, 400), V_th=-50., V_reset=-60.,
V_rest=-70., tau=10., tau_ref=2., noise=3. / bm.sqrt(10.),
V_initializer=bp.init.Uniform(-70., -50.),
method='exp_auto',
keep_size=True,
ref_var=True)
# delays
self.intra_delay_step = int(2. / bm.get_dt())
self.E_delay_steps = bm.asarray(delay_mat.T / bm.get_dt(), dtype=int)
bm.fill_diagonal(self.E_delay_steps, self.intra_delay_step)
self.Edelay = bm.LengthDelay(self.E.spike, delay_len=int(self.E_delay_steps.max()))
self.Idelay = bm.LengthDelay(self.I.spike, delay_len=self.intra_delay_step)
# synapse model
self.f_EE_current = partial(multiple_area_customized_op_E, conn_prob=0.1, shape=(1600, 1600), area_num=num_area)
self.f_EI_current = partial(multiple_area_customized_op_E, conn_prob=0.1, shape=(1600, 400), area_num=num_area)
self.f_IE_current = partial(multiple_area_customized_op_I, conn_prob=0.1, shape=(400, 1600), area_num=num_area)
self.f_II_current = partial(multiple_area_customized_op_I, conn_prob=0.1, shape=(400, 400), area_num=num_area)
# synapses from I
# self.intra_I2E_conn = bm.random.random((num_area, 400, 1600)) < 0.1
# self.intra_I2I_conn = bm.random.random((num_area, 400, 400)) < 0.1
self.intra_I2E_weight = -wEI
self.intra_I2I_weight = -wII
# synapses from E
# self.E2E_conns = [bm.random.random((num_area, 1600, 1600)) < 0.1 for _ in range(num_area)]
# self.E2I_conns = [bm.random.random((num_area, 1600, 400)) < 0.1 for _ in range(num_area)]
self.E2E_weights = (1 + alpha * hier) * muEE * conn.T # inter-area connections
bm.fill_diagonal(self.E2E_weights, (1 + alpha * hier) * wEE) # intra-area connections
self.E2I_weights = (1 + alpha * hier) * muIE * conn.T # inter-area connections
bm.fill_diagonal(self.E2I_weights, (1 + alpha * hier) * wIE) # intra-area connections
self.E_seeds = bm.random.randint(0, 100000, (num_area, num_area * 2))
self.I_seeds = bm.random.randint(0, 100000, (num_area * 2))
def update(self, v1_input):
self.E.input[0] += v1_input
self.E.input += self.extE
self.I.input += self.extI
E_not_ref = bm.logical_not(self.E.refractory)
I_not_ref = bm.logical_not(self.I.refractory)
# synapses from E
for i in range(self.num_area):
delayed_E_spikes = self.Edelay(self.E_delay_steps[i], i).astype(float)[i]
current = self.f_EE_current(delayed_E_spikes, self.E2E_weights[i], self.E_seeds[i, :self.num_area])
self.E.V += current * E_not_ref # E2E
current = self.f_EI_current(delayed_E_spikes, self.E2I_weights[i], self.E_seeds[i, :self.num_area])
self.I.V += current * I_not_ref # E2I
# synapses from I
delayed_I_spikes = self.Idelay(self.intra_delay_step).astype(float)
current = self.f_IE_current(delayed_I_spikes, self.intra_I2E_weight, self.I_seeds[:self.num_area])
self.E.V += current * E_not_ref # I2E
current = self.f_II_current(delayed_I_spikes, self.intra_I2I_weight, self.I_seeds[:self.num_area])
self.I.V += current * I_not_ref # I2I
# updates
self.Edelay.update(self.E.spike)
self.Idelay.update(self.I.spike)
self.E.update()
self.I.update()
Load data from .mat file
[30]:
# hierarchy values
hierVals = loadmat('Joglekar_2018_data/hierValspython.mat')
hierValsnew = hierVals['hierVals'].flatten()
hier = bm.asarray(hierValsnew / max(hierValsnew)) # hierarchy normalized.
# fraction of labeled neurons
flnMatp = loadmat('Joglekar_2018_data/efelenMatpython.mat')
conn = bm.asarray(flnMatp['flnMatpython'].squeeze()) # fln values..Cij is strength from j to i
# Distance
speed = 3.5 # axonal conduction velocity
distMatp = loadmat('Joglekar_2018_data/subgraphWiring29.mat')
distMat = distMatp['wiring'].squeeze() # distances between areas values..
delayMat = bm.asarray(distMat / speed)
Define parameters and run it
[31]:
pars = dict(extE=14.2, extI=14.7, wII=.075, wEE=.01, wIE=.075, wEI=.0375, muEE=.0375, muIE=0.0475)
inps = dict(value=15, duration=150)
inputs, length = bp.inputs.section_input(values=[0, inps['value'], 0.],
durations=[300., inps['duration'], 500],
return_length=True)
net = MultiAreaNet(hier, conn, delayMat, **pars)
runner = bp.DSRunner(net, monitors={'E.spike': lambda : net.E.spike.flatten()})
runner.run(inputs=inputs)
Predict 9500 steps: : 100%|██████████| 9500/9500 [01:32<00:00, 102.41it/s]
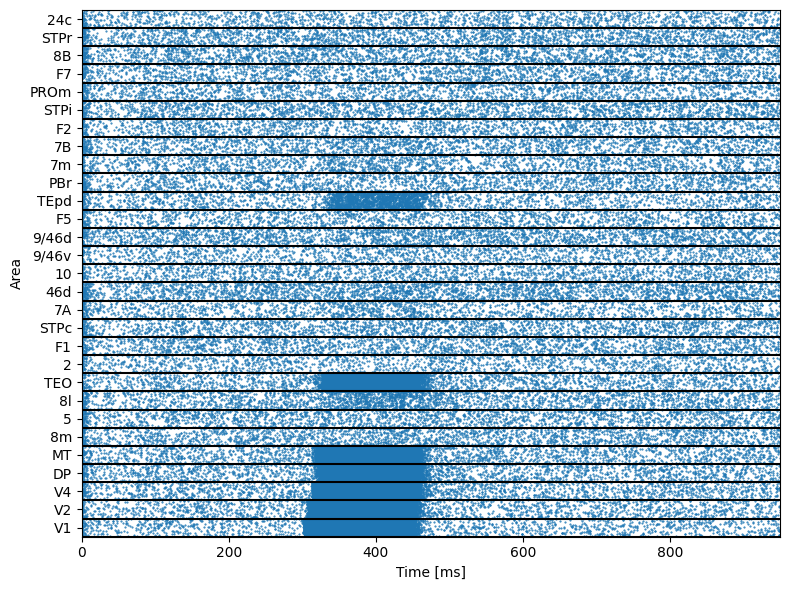
Draw the raster plot
[32]:
def raster_plot(xValues, yValues, duration):
ticks = np.round(np.arange(0, 29) + 0.5, 2)
areas = ['V1', 'V2', 'V4', 'DP', 'MT', '8m', '5', '8l', 'TEO', '2', 'F1',
'STPc', '7A', '46d', '10', '9/46v', '9/46d', 'F5', 'TEpd', 'PBr',
'7m', '7B', 'F2', 'STPi', 'PROm', 'F7', '8B', 'STPr', '24c']
N = len(ticks)
plt.figure(figsize=(8, 6))
plt.plot(xValues, yValues / (4 * 400), '.', markersize=1)
plt.plot([0, duration], np.arange(N + 1).repeat(2).reshape(-1, 2).T, 'k-')
plt.ylabel('Area')
plt.yticks(np.arange(N))
plt.xlabel('Time [ms]')
plt.ylim(0, N)
plt.yticks(ticks, areas)
plt.xlim(0, duration)
plt.tight_layout()
plt.show()
times, indices = np.where(runner.mon['E.spike'])
times = runner.mon.ts[times]
raster_plot(times, indices, length)