(Song, et al., 2016): Training excitatory-inhibitory recurrent network
Implementation of the paper:
Song, H. F. , G. R. Yang , and X. J. Wang . “Training Excitatory-Inhibitory Recurrent Neural Networks for Cognitive Tasks: A Simple and Flexible Framework.” Plos Computational Biology 12.2(2016):e1004792.
The original code is based on PyTorch (https://github.com/gyyang/nn-brain/blob/master/EI_RNN.ipynb). However, comparing with the PyTorch codes, the training on BrainPy speeds up nearly four folds.
Here we will train recurrent neural network with excitatory and inhibitory neurons on a simple perceptual decision making task.
[1]:
import brainpy as bp
bp.set_platform('cpu')
bp.math.use_backend('jax')
from brainpy.math import jax as bm
from brainpy.simulation import layers
[2]:
import numpy as np
import matplotlib.pyplot as plt
Defining a perceptual decision making task
[3]:
# We will import the task from the neurogym library.
# Please install neurogym:
#
# https://github.com/neurogym/neurogym
import neurogym as ngym
[4]:
# Environment
task = 'PerceptualDecisionMaking-v0'
timing = {
'fixation': ('choice', (50, 100, 200, 400)),
'stimulus': ('choice', (100, 200, 400, 800)),
}
kwargs = {'dt': 20, 'timing': timing}
seq_len = 100
# Make supervised dataset
dataset = ngym.Dataset(task,
env_kwargs=kwargs,
batch_size=16,
seq_len=seq_len)
# A sample environment from dataset
env = dataset.env
# Visualize the environment with 2 sample trials
_ = ngym.utils.plot_env(env, num_trials=2, fig_kwargs={'figsize': (10, 6)})
plt.show()
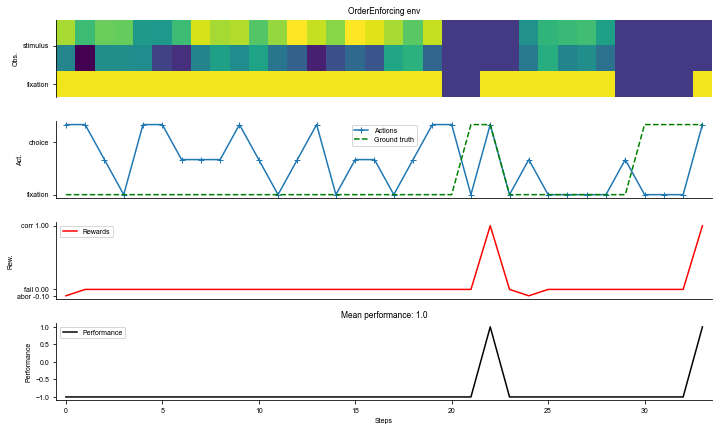
[4]:
input_size = env.observation_space.shape[0]
output_size = env.action_space.n
batch_size = dataset.batch_size
print(f'Input size = {input_size}')
print(f'Output size = {output_size}')
print(f'Bacth size = {batch_size}')
Input size = 3
Output size = 3
Bacth size = 16
Define E-I recurrent network
Here we define a E-I recurrent network, in particular, no self-connections are allowed.
[5]:
class RNN(layers.Module):
r"""E-I RNN.
The RNNs are described by the equations
.. math::
\begin{gathered}
\tau \dot{\mathbf{x}}=-\mathbf{x}+W^{\mathrm{rec}} \mathbf{r}+W^{\mathrm{in}}
\mathbf{u}+\sqrt{2 \tau \sigma_{\mathrm{rec}}^{2}} \xi \\
\mathbf{r}=[\mathbf{x}]_{+} \\
\mathbf{z}=W^{\text {out }} \mathbf{r}
\end{gathered}
In practice, the continuous-time dynamics are discretized to Euler form
in time steps of size :math:`\Delta t` as
.. math::
\begin{gathered}
\mathbf{x}_{t}=(1-\alpha) \mathbf{x}_{t-1}+\alpha\left(W^{\mathrm{rec}} \mathbf{r}_{t-1}+
W^{\mathrm{in}} \mathbf{u}_{t}\right)+\sqrt{2 \alpha \sigma_{\mathrm{rec}}^{2}} \mathbf{N}(0,1) \\
\mathbf{r}_{t}=\left[\mathbf{x}_{t}\right]_{+} \\
\mathbf{z}_{t}=W^{\mathrm{out}} \mathbf{r}_{t}
\end{gathered}
where :math:`\alpha = \Delta t/\tau` and :math:`N(0, 1)` are normally distributed
random numbers with zero mean and unit variance, sampled independently at every time step.
"""
def __init__(self, num_input, num_hidden, num_output, num_batch,
dt=None, e_ratio=0.8, sigma_rec=0., seed=None,
w_ir=bp.init.KaimingUniform(scale=1.),
w_rr=bp.init.KaimingUniform(scale=1.),
w_ro=bp.init.KaimingUniform(scale=1.)):
super(RNN, self).__init__()
# parameters
self.tau = 100
self.num_batch = num_batch
self.num_input = num_input
self.num_hidden = num_hidden
self.num_output = num_output
self.e_size = int(num_hidden * e_ratio)
self.i_size = num_hidden - self.e_size
if dt is None:
self.alpha = 1
else:
self.alpha = dt / self.tau
self.sigma_rec = (2 * self.alpha) ** 0.5 * sigma_rec # Recurrent noise
self.rng = bm.random.RandomState(seed=seed)
# hidden mask
mask = np.tile([1] * self.e_size + [-1] * self.i_size, (num_hidden, 1))
np.fill_diagonal(mask, 0)
self.mask = bm.asarray(mask, dtype=bm.float_)
# input weight
self.w_ir = self.get_param(w_ir, (num_input, num_hidden))
# recurrent weight
bound = 1 / num_hidden ** 0.5
self.w_rr = self.get_param(w_rr, (num_hidden, num_hidden))
self.w_rr[:, :self.e_size] /= (self.e_size / self.i_size)
self.b_rr = bm.TrainVar(self.rng.uniform(-bound, bound, num_hidden))
# readout weight
bound = 1 / self.e_size ** 0.5
self.w_ro = self.get_param(w_ro, (self.e_size, num_output))
self.b_ro = bm.TrainVar(self.rng.uniform(-bound, bound, num_output))
# variables
self.h = bm.Variable(bm.zeros((num_batch, num_hidden)))
self.o = bm.Variable(bm.zeros((num_batch, num_output)))
def cell(self, x, h):
ins = x @ self.w_ir + h @ (bm.abs(self.w_rr) * self.mask) + self.b_rr
state = h * (1 - self.alpha) + ins * self.alpha
state += self.sigma_rec * self.rng.randn(self.num_hidden)
return bm.relu(state)
def readout(self, h):
return h @ self.w_ro + self.b_ro
def make_update(self, h: bm.JaxArray, o: bm.JaxArray):
def f(x):
h.value = self.cell(x, h.value)
o.value = self.readout(h.value[:, :self.e_size])
return f
def predict(self, xs):
self.h[:] = 0.
f = bm.make_loop(self.make_update(self.h, self.o),
dyn_vars=self.vars(),
out_vars=[self.h, self.o])
return f(xs)
def loss(self, xs, ys):
hs, os = self.predict(xs)
os = os.reshape((-1, os.shape[-1]))
return bm.losses.cross_entropy_loss(os, ys.flatten())
Train the network on the decision making task
[6]:
# Instantiate the network and print information
hidden_size = 50
net = RNN(num_input=input_size,
num_hidden=hidden_size,
num_output=output_size,
num_batch=batch_size,
dt=env.dt,
sigma_rec=0.15)
[7]:
# Adam optimizer
opt = bm.optimizers.Adam(lr=0.001, train_vars=net.train_vars().unique())
[8]:
# gradient function
grad_f = bm.grad(net.loss,
dyn_vars=net.vars(),
grad_vars=net.train_vars().unique(),
return_value=True)
[9]:
@bm.jit
@bm.function(nodes=(net, opt))
def train(xs, ys):
grads, loss = grad_f(xs, ys)
opt.update(grads)
return loss
The training speeds up nearly 4 times, comparing with the original PyTorch codes.
[10]:
running_loss = 0
print_step = 200
for i in range(5000):
inputs, labels = dataset()
inputs = bm.asarray(inputs)
labels = bm.asarray(labels)
loss = train(inputs, labels)
running_loss += loss
if i % print_step == (print_step - 1):
running_loss /= print_step
print('Step {}, Loss {:0.4f}'.format(i + 1, running_loss))
running_loss = 0
Step 200, Loss 0.5044
Step 400, Loss 0.3405
Step 600, Loss 0.2425
Step 800, Loss 0.1826
Step 1000, Loss 0.1420
Step 1200, Loss 0.1171
Step 1400, Loss 0.1033
Step 1600, Loss 0.0928
Step 1800, Loss 0.0866
Step 2000, Loss 0.0800
Step 2200, Loss 0.0792
Step 2400, Loss 0.0739
Step 2600, Loss 0.0739
Step 2800, Loss 0.0709
Step 3000, Loss 0.0694
Step 3200, Loss 0.0680
Step 3400, Loss 0.0674
Step 3600, Loss 0.0660
Step 3800, Loss 0.0658
Step 4000, Loss 0.0678
Step 4200, Loss 0.0638
Step 4400, Loss 0.0641
Step 4600, Loss 0.0649
Step 4800, Loss 0.0636
Step 5000, Loss 0.0633
Run the network post-training and record neural activity
[11]:
predict = bm.jit(net.predict, dyn_vars=net.vars())
[12]:
env.reset(no_step=True)
env.timing.update({'fixation': ('constant', 500), 'stimulus': ('constant', 500)})
perf = 0
num_trial = 500
activity_dict = {}
trial_infos = {}
stim_activity = [[], []] # response for ground-truth 0 and 1
for i in range(num_trial):
env.new_trial()
ob, gt = env.ob, env.gt
inputs = bm.asarray(ob[:, np.newaxis, :])
rnn_activity, action_pred = predict(inputs)
# Compute performance
action_pred = action_pred.numpy()
choice = np.argmax(action_pred[-1, 0, :])
correct = choice == gt[-1]
# Log trial info
trial_info = env.trial
trial_info.update({'correct': correct, 'choice': choice})
trial_infos[i] = trial_info
# Log stimulus period activity
rnn_activity = rnn_activity.numpy()[:, 0, :]
activity_dict[i] = rnn_activity
# Compute stimulus selectivity for all units
# Compute each neuron's response in trials where ground_truth=0 and 1 respectively
rnn_activity = rnn_activity[env.start_ind['stimulus']: env.end_ind['stimulus']]
stim_activity[env.trial['ground_truth']].append(rnn_activity)
print('Average performance', np.mean([val['correct'] for val in trial_infos.values()]))
Average performance 0.846
Plot neural activity from sample trials
[13]:
trial = 2
plt.figure(figsize=(8, 6))
_ = plt.plot(activity_dict[trial][:, :net.e_size], color='blue', label='Excitatory')
_ = plt.plot(activity_dict[trial][:, net.e_size:], color='red', label='Inhibitory')
plt.xlabel('Time step')
plt.ylabel('Activity')
plt.show()
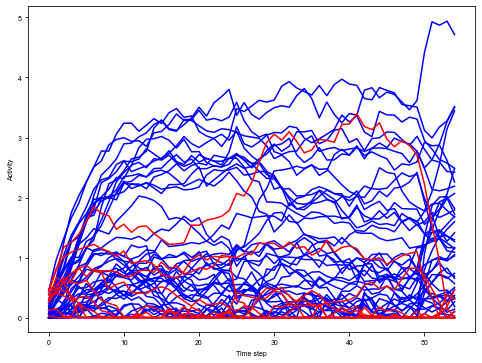
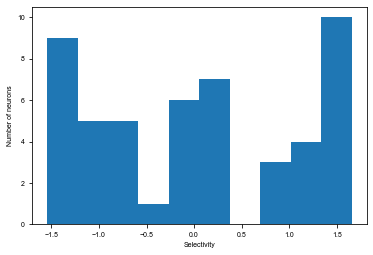
Compute stimulus selectivity for sorting neurons
Here for each neuron we compute its stimulus period selectivity \(d'\)
[14]:
mean_activity = []
std_activity = []
for ground_truth in [0, 1]:
activity = np.concatenate(stim_activity[ground_truth], axis=0)
mean_activity.append(np.mean(activity, axis=0))
std_activity.append(np.std(activity, axis=0))
# Compute d'
selectivity = (mean_activity[0] - mean_activity[1])
selectivity /= np.sqrt((std_activity[0] ** 2 + std_activity[1] ** 2 + 1e-7) / 2)
# Sort index for selectivity, separately for E and I
ind_sort = np.concatenate((np.argsort(selectivity[:net.e_size]),
np.argsort(selectivity[net.e_size:]) + net.e_size))
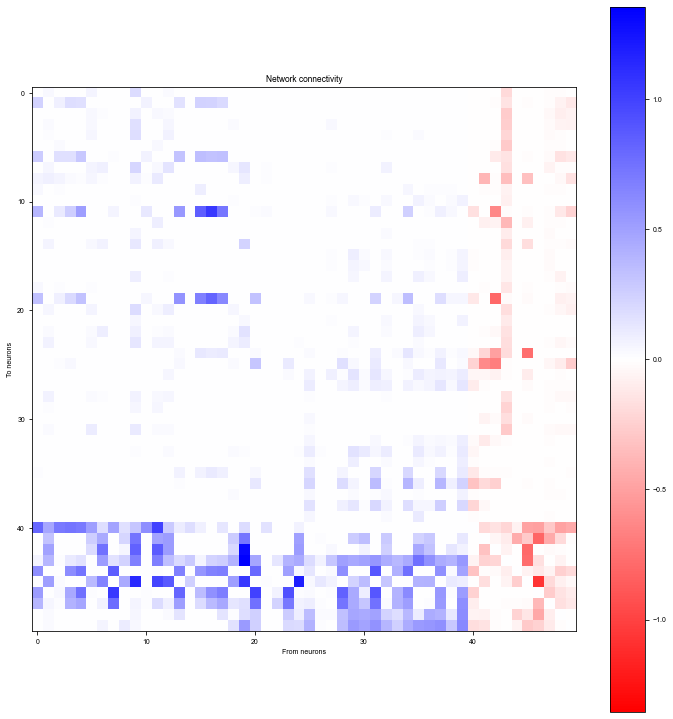
Plot network connectivity sorted by stimulus selectivity
[15]:
# Plot distribution of stimulus selectivity
plt.figure(figsize=(6, 4))
plt.hist(selectivity)
plt.xlabel('Selectivity')
plt.ylabel('Number of neurons')
plt.show()
[16]:
W = (bm.abs(net.w_rr) * net.mask).numpy()
# Sort by selectivity
W = W[:, ind_sort][ind_sort, :]
wlim = np.max(np.abs(W))
plt.figure(figsize=(10, 10))
plt.imshow(W, cmap='bwr_r', vmin=-wlim, vmax=wlim)
plt.colorbar()
plt.xlabel('From neurons')
plt.ylabel('To neurons')
plt.title('Network connectivity')
plt.tight_layout()
plt.show()